Evidence Review of the Adverse Effects of COVID-19 Vaccination and Intramuscular Vaccine Administration (2024)
Chapter: 2 Immunologic Response to COVID-19 Vaccines
2
Immunologic Response to COVID-19 Vaccines
The global pandemic stemming from the emergence of the novel severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) virus in late 2019 made it critical to develop efficacious vaccines. This public health crisis initiated global efforts to produce vaccines to reduce viral transmission and protect individuals from life-threatening infections (Diamond and Pierson, 2020). Several COVID-19 vaccines were rapidly developed using a variety of platforms. Concomitant with the release of vaccines, concerns arose around vaccine-induced harms. To better understand how vaccine-mediated harms may arise, it is important to know how specific COVID-19 vaccines initiate an immune response.
Charged with examining biological mechanisms, the committee conducted a comprehensive review of the current literature, examining the available evidence encompassing clinical trials, epidemiology studies, case reports, preclinical and translational in vitro or in silico studies, and insights gained from animal models. The committee analyzed a diverse array of vaccine-mediated harms and a variety of vaccine platforms and compiled a list of mechanisms that were deemed most plausible in contributing to the emergence of vaccine-mediated adverse reactions following COVID-19 vaccination. Throughout these deliberations, the committee engaged in in-depth discussions regarding the pathophysiology that may be involved and the requisite evidentiary support necessary to establish the presence of a particular mechanism.
FUNDAMENTALS OF THE IMMUNE RESPONSE
The human immune response is initiated by the innate immune system, which activates the adaptive immune system. Both the innate and adaptive arms of the immune response play a pivotal role in combating pathogens, such as SARS-CoV-2, and establishing long-term immunity. They are also both important in producing an effective immune response and long-term immunity (immunological memory) after vaccination.
The innate immune system is the “first responder” to foreign agents, such as viral infections or physical tissue damage. It comprises physical defenses, such as the skin, and cellular components, such as macrophages, mast cells, dendritic cells, neutrophils, and natural killer cells. The innate immune response is not pathogen specific at the single amino acid/protein epitope level (i.e., antigen) but recognizes categories of pathogens, such as viruses, bacteria, parasites, and tissue damage, based on molecular patterns that are specific to particular microbes (Chaplin, 2010). Important to this pathogen recognition system are pattern recognition receptors, with Toll-like receptors (TLRs) being a notable subgroup. For example, TLR3 is involved in canonically recognizing double-stranded ribonucleic
acid (RNA), commonly associated with viral infections; however, evidence of TLR3 recognition of single-stranded RNA vaccines has been shown (Teijaro and Farber, 2021). TLR4, which recognizes lipopolysaccharides from Gram-negative bacteria, some viral infections, and self-like ATP from damaged mitochondria, may play a role in responses to messenger ribonucleic acid (mRNA) vaccines, which contain mRNA within lipid nanoparticles (LNPs) that augment innate immune responses. TLR7 and TLR8, recognizing single-stranded RNA, are integral to the immune response against RNA-based vaccines, such as certain COVID-19 vaccines. Meanwhile, TLR9, which detects unmethylated CpG motifs in bacterial and viral deoxyribonucleic acid (DNA), is used in some vaccines as an adjuvant (a substance in vaccines that enhances the immunological response to the antigen). The activation of these TLRs triggers signaling pathways that lead to cytokine and type I interferon production, crucial for initiating adaptive immune responses (Fitzgerald and Kagan, 2020).
After activation by the innate immune system, the adaptive immune system develops an antigen-specific immune response to a specific pathogen that is based on particular amino acids/protein sequences (antigens). Macrophages and dendritic cells are fundamental in presenting antigens to adaptive immune cells, initiating an antigen-specific response crucial for establishing long-lasting immunological memory. Because of the high specificity of the adaptive immune response, it can distinguish not only a specific virus but also a specific strain of that virus. Memory occurs primarily at the T cell and B cell levels. B cells develop into plasma cells that release antigen-specific antibodies that are critical for rapidly clearing infections when they are encountered the next time. T cells, on the other hand, play a crucial role in immune memory by recognizing and responding to previously encountered antigens, aiding in the rapid mobilization of the immune system during subsequent infections. The development of strong antigen-specific T cell and B cell-antibody memory is a primary goal of vaccine development.
All types of vaccines strongly stimulate an innate immune response to direct the adaptive immune response to make protective antigen-specific T and B cells and antibody responses against the target infection. COVID-19 vaccines (see Figure 2-1), including traditional protein-based vaccines (NVX-CoV23731), mRNA vaccines (e.g., BNT162b22 and mRNA-12733) and adenovirus-vector (AV) vaccines (e.g., Ad26.COV2.S4 and ChAdOx1/nCoV-195), are engineered to stimulate both innate and adaptive immune responses. The mRNA vaccines deliver genetic material coding for the SARS-CoV-2 spike S-protein into host cells (Martinez-Flores et al., 2021) so that an antigen-specific adaptive immune response will be generated against it. The mRNA vaccine may be able to activate resident innate immune cells at the injection site, but it primarily takes effect after the spike protein is generated within cells (Verbeke et al., 2022). The mRNA strands are structurally optimized to prevent degradation by incorporating pseudouridines (Kim et al., 2022) and mRNA into LNPs (Ndeupen et al., 2021), which both further protects the RNA transcript from degradation and facilitates cell entry (Pardi et al., 2015). Certain components within the LNP layer may also act as adjuvants by activating TLRs on antigen presenting cells and the innate immune response to induce an enhanced adaptive immune response against the spike protein (Alameh et al., 2021). Protein-based vaccines often require an adjuvant to stimulate the innate immune response; AV vaccines have an innate immune-activating ability because they are viral vectors.
Innate antigen presenting cells, particularly mast cells, macrophages, and dendritic cells, are instrumental in activating an adaptive immune response. They capture, process, and present pathogen-specific antigens to T cells, inducing a highly targeted adaptive response and immunological memory. Dendritic cells are particularly important in stimulating adaptive immune responses from the draining lymph nodes while resident mast cells and macrophages play key roles at tissue sites. In the milieu of COVID-19 vaccines, antigen presenting cells are vital for identifying and presenting the vaccine-derived spike protein to helper T cells, thereby producing spike protein–specific T and B cells.
T cells, comprising helper T cells (CD4+) and cytotoxic T cells (CD8+), play multifaceted effector roles during an infection such as SARS-CoV-2. Helper T cells facilitate B cell activation and enhance the function of cytotoxic T cells, which directly attack and destroy virally infected cells. The adaptive immune cell memory induced by
___________________
1 Refers to the COVID-19 vaccine manufactured by Novavax.
2 Refers to the COVID-19 vaccine manufactured by Pfizer-BioNTech under the name Comirnaty®.
3 Refers to the COVID-19 vaccine manufactured by Moderna under the name Spikevax®.
4 Refers to the COVID-19 vaccine manufactured by Janssen.
5 Refers to the COVID-19 vaccine manufactured by Oxford-AstraZeneca.
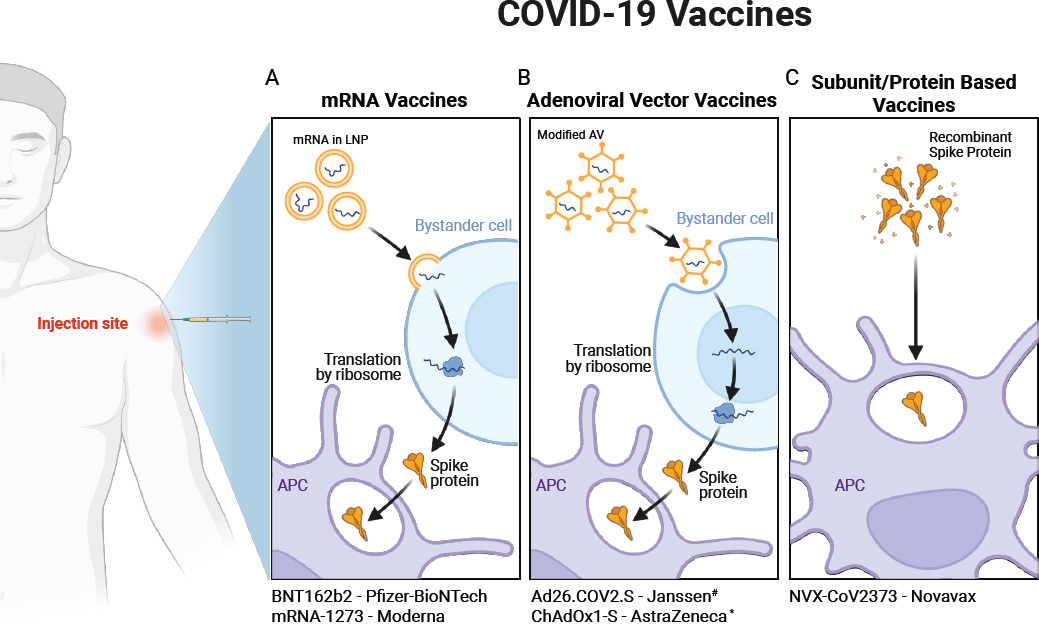
NOTES: (A) mRNA Vaccines: Upon injection, mRNA encapsulated in lipid nanoparticles (LNPs) is delivered into myocytes or bystander cells. The mRNA is released from LNPs and translated by ribosomes to produce the viral antigen, such as the spike protein (S), which is secreted. Antigen-presenting cells (APCs) such as dendritic cells (DCs) uptake the secreted antigen, initiating an immune response. (B) Adenoviral Vector Vaccines: Adenoviral vectors containing viral DNA enter myocytes or bystander cells, where they uncoat. The DNA, containing a nuclear localization signal, is transported to the nucleus and transcribed into mRNA. The extrachromosomal DNA does not integrate into the host genome. The mRNA is translated into protein, which is secreted and uptaken by APCs, initiating an immune response. (C) Subunit Vaccines: Pre-formed viral protein, such as the spike protein (S), is delivered. Antigen-presenting cells, particularly resident dendritic cells (DCs), uptake the protein to initiate an immune response. Additionally, M-matrix adjuvants enhance this response. #Ad26.COV2.S is no longer authorized under EUA in the United States as of June 1, 2023. *ChAdOx1-S is not used in the United States. Created with BioRender.com.
COVID-19 vaccines ensures a rapid antigen-specific T and B cell/antibody response when the vaccinee encounters SARS-CoV-2 in the future.
SARS-COV-2 AND VACCINE TARGET OF THE SPIKE PROTEIN
SARS-CoV-2 is characterized by several structural proteins; the spike (S) glycoprotein and the nucleocapsid (N) protein are primary targets for the immune response (Krammer, 2020). The spike protein is a major virus surface protein crucial for viral entry into host cells; it binds to the angiotensin-converting enzyme 2 (ACE2) receptor on host cells (Walls et al., 2020). Structurally, the spike protein is a class I viral fusion glycoprotein comprising of two subunits: the S1 subunit, responsible for receptor binding, and the S2 subunit, involved in fusion. These subunits are connected by a furin cleavage site, unique to SARS-CoV-2 (rather than all SARS viruses), and the protein is cleaved posttranslationally at this furin cleavage site. The receptor-binding domain (RBD) within the
S1 subunit is particularly critical for viral entry to cells, as it directly interacts with the ACE2 receptor, initiating conformational changes leading to membrane fusion and viral entry (Kirchdoerfer et al., 2016; Wrapp et al., 2020). In addition, the spike protein is the only SARS-CoV-2 antigen recognized to stimulate neutralizing antibodies (Xiaojie et al., 2020). Several other receptors are important in viral entry but not described in this report.
The spike protein has been the primary focus in vaccine developments due to its essential role in viral entry to host cells. Vaccines contain (subunit vaccines, such as NVX-CoV2373) or generate production of (mRNA and AV vaccines) the spike protein to elicit an immune response in the absence of infection. Typically, adjuvants are also needed to induce a strong immune response because the antigen itself (without an active infection) does not do so in individuals who have not encountered SARS-CoV-2. The goal of all vaccine platforms is to contain or produce a stable form of the S protein that will not degrade or be cleared from the body without activating the immune response.
Two main mRNA vaccine strategies have been employed to stabilize the spike protein in its prefusion conformation, which is essential for preserving epitopes that are sensitive to degradation. One method, used in both mRNA-1273 and BNT162b2 vaccines, introduces mutations in the mRNA transcript (proline substitutions at positions 986 and 987), which maintain the spike glycoprotein in the prefusion state (Pallesen et al., 2017; Wrapp et al., 2020). Another strategy, not employed by the current vaccines, involves designing an mRNA construct where the full-length spike protein lacks the furin cleavage site (∆furin), preventing posttranslational cleavage (Laczko et al., 2020; Lederer et al., 2020).
As an alternative to targeting the full-length spike protein, some vaccines focus solely on RBD (Bettini and Locci, 2021), which contains multiple epitopes that can be effective targets for virus neutralization, making it a potent target for vaccine strategies (Robbiani et al., 2020; Zost et al., 2020). For instance, the BNT162b1 vaccine candidate developed by BioNTech/Pfizer encodes a secreted trimerized version of RBD. The choice of the full-length spike protein or smaller RBD of the spike protein in vaccine design balances the benefits of eliciting a broader immune response with the full-length protein versus focusing on the highly neutralizing epitopes in the RBD. However, due to its favorable immunogenicity to reactogenicity profiles, BNT162b2, encoding full-length spike protein, was chosen as the leading vaccine candidate (Khehra et al., 2021).
TYPES OF COVID-19 VACCINES
Several COVID-19 vaccines have been developed and authorized for use in the United States, using several different vaccine platforms (see Figure 2-1). The mRNA vaccines, such as BNT162b2 and mRNA-1273, use LNP-encapsulated mRNA to encode the SARS-CoV-2 spike protein. This technology prompts host cells to produce the spike protein, subsequently eliciting innate and adaptive immune responses and, most importantly, immunological memory. Adenovirus vector vaccines, such as Ad26.COV2.S (Emergency Use Authorization was revoked by the Food and Drug Administration [FDA] on June 1, 2023) and AZD1222 (not used in the United States), employ modified adenoviruses to deliver DNA encoding the spike protein. Protein subunit vaccines, such as NVX-CoV2373, consist of recombinantly produced viral proteins (such as the spike protein or its epitopes) combined with the Matrix-M® adjuvant, which enhances the immunogenicity of the protein antigen, leading to a more robust immune response. Each platform has distinct immunogenic profiles and mechanisms for eliciting an immune response.
mRNA Vaccines
The advent of mRNA vaccines has marked a revolutionary leap in the field of immunology and vaccine development, particularly underscored by their critical role in combating the COVID-19 pandemic. These vaccines represent a significant departure from traditional vaccine platforms, providing a number of new advantages, including rapid development, high efficacy and safety, and rapid adaptation to new viral strains (Welsh, 2021). This technology holds promise for preventing serious outcomes and/or spread from viral infections. Developing mRNA vaccines, although conceptually straightforward, involves a complex design process. These vaccines function by delivering mRNA encoding a target antigen, such as the SARS-CoV-2 spike protein, into host cells. Once in the cytoplasm, the cells use their own machinery to translate the mRNA into the target protein, which is released from
the host cell, usually in an extracellular vesicle (Trougakos et al., 2022), activating an innate immune response that also prompts the adaptive immune system to mount a memory response against the spike (S) protein. The next time the individual sees the spike protein during an active infection or vaccine boost, the immune system rapidly mounts a highly protective T and B cell/antibody response. For a detailed depiction of the sequence through which SARS-CoV-2 mRNA vaccines elicit immune responses, from their administration to the priming of T cells and initiation of germinal center (GC) reactions, refer to Figure 2-2.
One of the initial challenges of mRNA vaccines was the inherent nature of unmodified mRNA, which is extremely labile and highly immunogenic, making it unsuitable for direct use in vaccines (Pardi et al., 2018). Karikó et al. (2008) tested various modifications to nucleosides in mRNA molecules. They tested modifications, such as pseudouridine, 5-methylcytidine, N6-methyladenosine, 5-methyluridine, and 2-thiouridine. The substitution
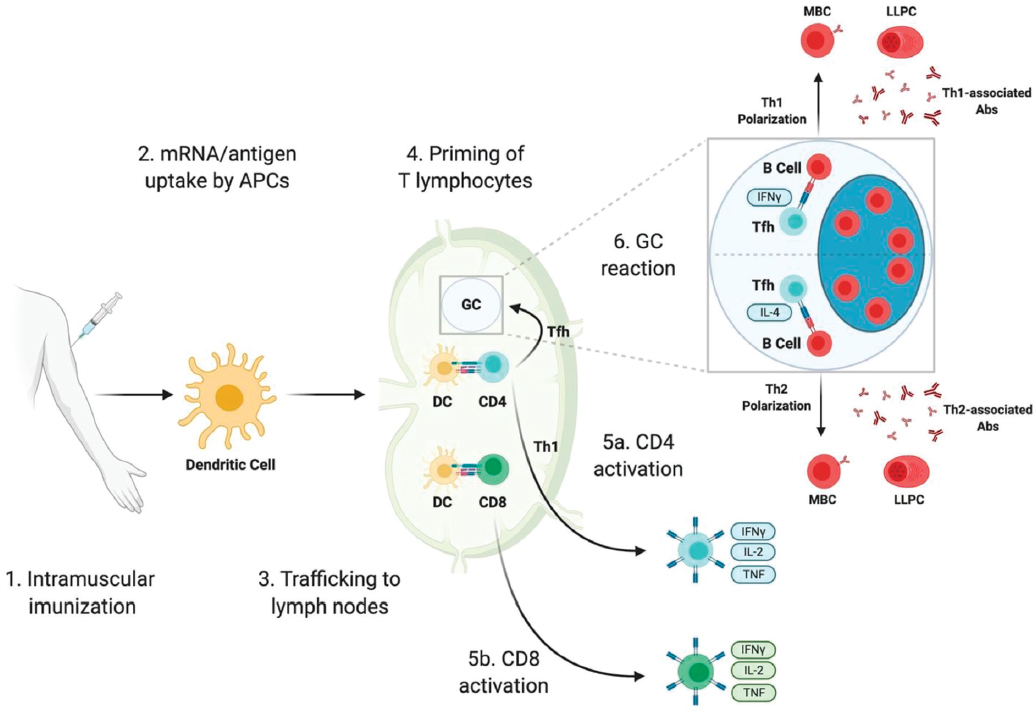
NOTES: Immune responses triggered by SARS-CoV-2 mRNA vaccines involve a sequence of events starting with their intramuscular administration. These vaccines, which include mRNA encapsulated in lipid nanoparticles (mRNA-LNPs) or the antigen they produce, are first taken up by antigen-presenting cells (APCs) such as dendritic cells. After uptake, these APCs migrate to the lymph nodes, where they activate both CD4 and CD8 T lymphocytes. This process of T cell priming and its subsequent steps are discussed comprehensively in scientific literature. Following priming, CD8 T cells may differentiate into cytotoxic T lymphocytes capable of destroying virus-infected cells, while CD4 T cells may evolve into either Th1 cells or T follicular helper (Tfh) cells. Tfh cells are pivotal in initiating the germinal center reaction, a critical process for the development of high-affinity memory B cells and long-lived plasma cells that secrete antibodies. The direction of Tfh cell differentiation toward a Th1 or Th2 phenotype influences the isotype of antibodies produced by these plasma cells, affecting the body’s immune response to the vaccine.
SOURCE: Bettini and Locci, 2021 CC BY.
of uridine with N1-methyl-pseudouridine (m1Ψ) led to a 10-fold increase in translation efficiency compared to unmodified mRNA (Karikó et al., 2008). Moreover, mRNA with this modification was not recognized by the pathogen-associated molecular pattern (PAMP) sensing mechanisms, such as TLRs or retinoic acid-inducible gene I (RIG-I), thus avoiding excessive inflammation, RNA degradation, and potential harms (Karikó et al., 2008; Pardi et al., 2018). This m1Ψ modification has been adopted in the design of several mRNA vaccine candidates, including the widely used mRNA-1273 and BNT162b2 (Corbett et al., 2020; Walsh et al., 2020).
Although it is possible to inject naked mRNA directly for immunization, this approach is generally inefficient (Cao and Gao, 2021). For the mRNA to be translated into proteins in the host cell, it must penetrate the cell’s lipid membrane to reach the cellular ribosomes. To facilitate efficient protein translation, delivery methods that ensure the cytosolic localization of mRNA are essential. Although standard laboratory lipid encapsulation methods, such as lipofectamine, were effective in vitro, they were cytotoxic and less efficient in vivo (Cao and Gao, 2021; Karikó et al., 2008). The encapsulation of mRNA into LNPs significantly contributes to its stability and uptake by the cells; LNPs effectively transport mRNA within the body and, upon intramuscular injection, can be taken up by antigen-presenting cells at the injection site and in nearby lymph nodes, facilitating both innate and adaptive immune responses. Furthermore, LNPs provide protection against nuclease-mediated degradation of the mRNA. The composition of LNPs is often proprietary, but they are known to contain a mixture of ionizable cationic lipids, cholesterol, phospholipids, and polyethylene glycols (PEGs), which self-assemble into nanoparticles of approximately 100 nanometers in diameter to encapsulate the mRNA (Cullis and Hope, 2017; Maier et al., 2013). Many of these components are known to be immunogenic and can act as adjuvants to stimulate the innate immune response to the spike protein. In fact, the composition of the LNP can be tailored to enhance the immune system’s response to the vaccine by inducing robust T follicular helper (Tfh) cell and humoral responses, making LNPs not only a delivery vehicle but also an adjuvant-like component of mRNA vaccines (Alameh et al., 2021).
Adenovirus-Vector Vaccines
AV vaccines have emerged as key players in COVID-19 vaccine development, leveraging the unique properties of adenoviruses. These linear double-stranded DNA viruses, typically responsible for respiratory infections in children and adults, possess stable genes and efficient transduction capabilities (ability to transfer genetic materials), making them ideal vaccine vectors (Lukashev and Zamyatnin, 2016). Adenoviruses do not integrate into the host genome but remain in a non-genome episomal state, meaning the injected genetic material translocates into the nucleus but does not integrate into the host DNA (Coughlan, 2020; Walsh et al., 2020). This aspect is significant because it mitigates concerns about potential long-term genetic changes in the host’s cells. In some other types of viral vectors, the viral DNA could integrate into the host’s genome, which could lead to unintended genetic alterations (Bulcha et al., 2021). However, with AV vaccines, this risk is greatly reduced because the adenovirus DNA remains separate from the host’s DNA.
The adenovirus’s nucleocapsid is composed of fiber, penton, and hexon proteins, contributing to its robustness and versatility as a vector. Over 150 primate adenoviruses have been identified, with many being developed for vaccines due to their cost-effectiveness, thermostability, and ability to induce strong immune responses (Chavda et al., 2023). A significant challenge is pre-existing immunity to common adenovirus serotypes in humans. To circumvent this, rare adenoviruses are employed, such as Ad26 or chimpanzee adenoviruses, which are less likely to be neutralized by pre-existing human antibodies. These vectors have demonstrated effectiveness in both animal models and human studies, despite the varying levels of pre-existing immunity across populations (Ewer et al., 2017; Geisbert et al., 2011).
In the context of COVID-19 vaccines, AZD1222, also known as “Covishield” by the Serum Institute of India, uses the ChAdOx1 AV. It carries the gene for the SARS-CoV-2 spike protein (ChAdOx1-S), which is expressed in its trimeric prefusion conformation (Watanabe et al., 2021).
Janssen Pharmaceuticals developed Ad26.COV2.S, using an Ad26 vector that encodes the spike protein with specific modifications (K986P and V987P) to enhance immunogenicity by locking the spike in its prefusion conformation (Bos et al., 2020). This vaccine is distinguished by its single-dose regimen.
Protein Subunit Vaccines
Protein-based vaccines, a well-established class of vaccines, use specific proteins (or protein fragments) from a pathogen to elicit an immune response without introducing the complete pathogen. These vaccines are known for their safety, as they do not contain live components of the pathogen, reducing the risk of vaccine-induced disease, but they are also less immunogenic and require adjuvants or other interventions (Pollard and Bijker, 2021). These vaccines fall into two main categories: subunit vaccines, which include only the parts of the virus that best stimulate the immune system, and toxoid vaccines, which use a toxin produced by the pathogen that has been made harmless but still triggers immunity. Toxoid vaccines include diphtheria and tetanus vaccines, which use inactivated forms of the toxins produced by these bacteria. Subunit vaccines include hepatitis B, which uses a surface protein from the virus, the pertussis toxin component of the DtaP (diphtheria, tetanus, and acellular pertussis) vaccine, and NVX-CoV2373.
NVX-CoV2373 comprises recombinantly produced spike proteins combined with Novavax’s proprietary Matrix-M® adjuvant (Keech et al., 2020). The spike protein used in the vaccine is produced by baculovirus expression in Spodoptera frugiperda insect cells, a method known for its ability to yield complex, properly folded proteins (Jarvis, 2003). This strategy ensures that the spike protein maintains its prefusion conformation, which is known to expose critical neutralizing epitopes more effectively than the post-fusion conformation (Bowen et al., 2021; Keech et al., 2020). The adjuvant is a critical component that significantly boosts the innate immune response to the spike protein. It is based on saponin, derived from the Quillaja saponaria tree, and combined with cholesterol and phospholipid to form nanoparticles. These nanoparticles enhance the immune response by stimulating the entry of the antigen into antigen-presenting cells and activating these innate cells. This adjuvant has been shown to boost both the quantity and quality of the immune response, leading to higher levels of neutralizing antibodies and a more robust T cell response (Stertman et al., 2023) to infection. In addition, adjuvants enable the use of smaller amounts of antigen. Producing neutralizing antibodies is the goal for most vaccines, as they bind to a pathogen and block its ability to infect cells, effectively neutralizing its disease-causing capabilities. In addition to antibody production, the orchestration of a robust T cell response is paramount, as these cells not only assist in the maturation of antibody-producing B cells but also identify and eliminate infected host cells, thereby mitigating the pathogen’s proliferation and ensuring a comprehensive immunological defense.
This vaccine’s storage and handling requirements are less stringent than those of mRNA vaccines, making it a valuable asset in global vaccination efforts, especially in regions with limited cold chain infrastructure.
VACCINE IMMUNE RESPONSE ELICITATION
For non-single-dose COVID-19 vaccines, the first and second doses play distinct and complementary roles in eliciting an effective immune response. The initial vaccine dose largely primes the immune system, providing the antigen in a way that stimulates initial antibody production and activates specific immune cells that lead to antigen-specific memory T and B cells. Because the vaccine is not an actual infection, it may not provide the needed cues to mount an optimal immune response. Thus, the second dose, or the booster, is crucial for amplifying and broadening this response. It significantly enhances the quantity and quality of neutralizing antibodies, solidifies memory B cell and T cell responses, and induces a more robust, durable immunity. The booster dose thus ensures a more sustained and effective immune response, including against virus variants (Chu et al., 2022). Table 2-1 presents a summary of antibody responses and T cell responses in humans for each U.S. COVID-19 vaccine.
The immunogenicity of COVID-19 vaccines largely hinges on the adaptive immune system recognizing the specific spike protein fragments. B cell receptors on B cells and T cell receptors on T cells are key to this recognition when they interact with innate immune antigen-presenting cells. B cell receptors directly bind to epitopes on the spike protein, initiating B cell activation (Pettini et al., 2022). T cell receptors, however, recognize these epitopes when presented on Major Histocompatibility Complex (MHC) molecules by antigen-presenting cells (Yang et al., 2023). This dual recognition mechanism is essential for the coordinated activation of both humoral and cellular arms of the adaptive immune response (Teijaro and Farber, 2021) for viral proteins that are not superantigens.
Following vaccination, B cell activation predominantly occurs in GCs within secondary lymphoid organs (see Figure 2-2), such as lymph nodes and the spleen. In general, antigen-activated B cells undergo somatic
TABLE 2-1 Immune Responses to U.S. COVID-19 Vaccines
| Vaccine | Platform | Dosing Regimen | Antibody Responses in Humans | T Cell Responses in Humans |
|---|---|---|---|---|
| BNT162b2 | mRNA | 30 μg mRNA 2 doses 21 days aparta | S1-binding antibody present after first dose, responses increased following the second dose; significant NAB was only present after second doseb | Increases in antigen-specific IFNγ+ CD4+ and CD8+ T cells after second dose; predominance of IFNγ and IL-2 secretion, compared with IL-4, suggesting TH1 cell polarizationc |
| mRNA-1273 | mRNA | 100 μg mRNA 2 doses 28 days apart | S-binding antibody detected 14 days after first dose, levels increased slightly by 28 days, with marked increase after second dosed; minimal NAB present after first dose, peak at 14 days after second dosee | Significant increases in CD4+ T cells secreting TH1 type cytokines (TNF > IL-2 > IFNγ) after second dose, small increases in TNF-secreting and IL-2secreting cells after first dose; minimal change in TH2 cell responses; low levels of CD8+ responsesd |
| Ad26.COV2.S | Viral vector | 5 × 1010 viral particles 1 dosef | S-binding and neutralizing antibody present by 28 days after vaccination in 99% of individuals and antibody levels sustained until at least 84 days post vaccinationg | CD4+ and CD8+ T cell responses present at 14 and 28 days post-vaccination, based on presence of CD4+ and CD8+ T cells secreting IFNγ and/or IL-2 and not IL-4 or IL-3, suggesting TH1 cell polarization of the CD4+ T cell responseg |
| NVX-CoV2373 | Protein subunit | 5 μg protein 2 doses 21 days aparth | S-binding antibody detected 21 days after first dose, with a marked increase after the second dose; some NAB present after the first dose, with a significant increase by 7 days after second doseh | CD4+ T cell responses present by 7 days after second dose, based on IFNγ, IL-2 and TNF production in response to S protein stimulation, with a strong bias toward a TH1 cell phenotype; minimal TH2 cell responses (as measured by IL-5 and IL-13)h |
NOTES: BNT162b2 refers to the COVID-19 vaccine manufactured by Pfizer-BioNTech under the name Comirnaty®. mRNA-1273 refers to the COVID-19 vaccine manufactured by Moderna under the name Spikevax®. Ad26.COV2.S refers to the COVID-19 vaccine manufactured by Janssen. NVX-CoV2373 refers to the COVID-19 vaccine manufactured by Novavax. CD: Cluster of Differentiation; IFN: Interferon; IL: Interleukin; mRNA: Messenger Ribonucleic Acid; NAB: Neutralizing Antibody; TH: T-helper cells; TNF: Tumor Necrosis Factor
c FDA, 2021; Sadarangani et al., 2021.
d FDA, 2022; Jackson et al., 2020.
SOURCE: Adapted from Sadarangani et al., 2021.
hypermutation, which introduces random mutations into their immunoglobulin (Ig) genes (Laidlaw and Ellebedy, 2022; Turner et al., 2021) and leads to B cells with high-affinity antibodies for the spike protein. B cells with the highest affinity are selected and differentiated into long-lived plasma cells (LLPCs) and memory B cells. LLPCs secrete neutralizing antibodies, some of which are capable of mediating sterilizing immunity, which prevents infection in the host including mucous membranes, and can potentially persist for years, continuously producing antibodies. Memory B cells quickly activate and give rise to a new wave of high-affinity antibody-secreting cells, providing rapid protection upon re-exposure to the virus (Sadarangani et al., 2021; Tam et al., 2016). For COVID-19 vaccines, in time, mutations in the spike protein may result in lower-affinity interaction between the antibodies induced by one strain and a mutated spike protein.
The role of T cells, particularly CD4+ T cells, is multifaceted. Tfh cells, a subset of CD4+ T cells, are critical for the development of GC reactions and consequently for the maturation of B cell responses. Tfh cells assist B cells in the GCs by providing necessary costimulatory signals and cytokines, facilitating the selection of high-affinity
B cells. These interactions are crucial for developing both LLPCs and memory B cells, and mRNA vaccines have been demonstrated to effectively induce Tfh cell responses, which are key to generating robust and long-lasting neutralizing immunity (Bettini and Locci, 2021; Pardi et al., 2018; Sadarangani et al., 2021). Clinically, however, immunity from COVID-19 vaccines is observed to wane over time (Menegale et al., 2023), necessitating booster doses to counteract this decline and to address the emergence of new, circulating common strains, thereby ensuring sustained protection against the virus.
Cytotoxic CD8+ T cells, which directly eliminate virus-infected cells, are another crucial component. These cells are characterized by the release of cytotoxic molecules, such as granzyme B and perforin. Upon vaccination, polyfunctional antigen-specific CD8+ T cells increase; these produce inflammatory cytokines, which are critical signaling molecules in the immune system. These include IFNγ (interferon gamma), IL-2 (interleukin-2), and TNF (tumor necrosis factor). IFNγ plays a crucial role in activating and directing other immune cells, enhancing the overall immune response to the vaccine and the virus. IL-2 is vital for the growth, proliferation, and differentiation of T cells, ensuring a robust and sustained immune response. TNF is involved in systemic inflammation and capable of inducing apoptosis or cell death in virus-infected cells. These cells exhibit markers of cytotoxic activity and contribute to the overall defense against viral infection. The ability to activate CD8+ T cell responses varies among vaccine candidates, with some inducing strong responses in both small and large animal models, while others show more variable results (Bettini and Locci, 2021; Creech et al., 2021).
Immunological memory is a hallmark of the adaptive immune response and a key goal of vaccination. Most licensed vaccines, including those for COVID-19, confer protection by eliciting long-lasting antibody responses.
The rapid and effective response to a pathogen upon re-exposure is primarily mediated by memory B and T cells. Memory B cells, upon re-exposure to the antigen, differentiate into antibody-secreting cells more quickly than naïve B cells, leading to a fast increase in antibody titers. Similarly, memory T cells, both CD4+ and CD8+, are primed to respond more rapidly and effectively than naïve T cells. Tfh cells are especially important in supporting memory B cell responses in GCs. They facilitate the selection of high-affinity memory B cells and their differentiation into LLPCs or memory B cells (Pollard and Bijker, 2021). These interactions are critical for maintaining long-lasting immunity and providing rapid protection upon subsequent exposures to the virus. Upon activation in a future infection, memory B cells rapidly produce large amounts of antigen-specific antibody, which can neutralize viral infection/entry into host cells—reducing the severity of the infection.
The duration of immunity conferred by COVID-19 vaccines and the potential need for booster doses are areas of ongoing research. Studies have shown that mRNA vaccines can induce robust CD8+ T cell responses characterized by key cytokines and cytotoxic markers upon rechallenge (Sadarangani et al., 2021; Teijaro and Farber, 2021). However, the longevity of these responses and persistence of memory T cells after vaccination is still under investigation. Some evidence suggests that the immune response elicited by these vaccines, particularly the generation of memory B and T cells, may be long lasting, but further studies are required to confirm the duration of this protection. Additionally, the need for booster doses may depend on factors such as the emergence of new viral variants/strains and the longevity of the vaccine-induced immune response (Teijaro and Farber, 2021).
POSSIBLE MECHANISMS OF VACCINE-MEDIATED REACTIONS
Although rare, vaccine-mediated harms can range from mild, transient reactions to more serious conditions, underscoring the importance of ongoing safety monitoring and research. Certain of the most common vaccine-associated harms can arise from a few different immunological mechanisms, some of which are briefly discussed next (see Table 2-2).
Immediate-type hypersensitivity reactions are rapid immunological responses observed in certain individuals following vaccination. Mast cells and basophils play a crucial role; they become activated by IgE when individuals are re-exposed to the same antigen during vaccination (Stone et al., 2019), which triggers degranulation and the release of various mediators, such as histamine, leukotrienes, prostaglandins, and cytokines, including IL-4 and IL-5. The clinical manifestation ranges from urticaria (hives) to the more severe and potentially life-threatening anaphylaxis (McLeod et al., 2015).
TABLE 2-2 Vaccine-Mediated Reactions and Their Mechanisms
| Type of Reaction | Immune Cells Involved | Plausible Mechanisms | Clinical Manifestation | Time of Onset |
|---|---|---|---|---|
| Immediate hypersensitivity | Mast cells, basophils | IgE-mediated mast cell/basophil activation induced by previous exposure to antigens in the vaccine, leading to degranulation and release of histamine, leukotrienes, prostaglandins, cytokines (IL-4, IL-5) | Urticaria (hives) to anaphylaxis | Rapid, post-vaccination |
| Delayed hypersensitivity | CD4+ helper T cells | Secretion of cytokines (IFN-γ, IL-2, TNF-α) upon activation | Rash, fever, joint pain | Days to weeks |
| Autoimmune reactions | Various | Molecular mimicry, bystander activation, epitope spreading, polyclonal activation, adjuvant-induced autoimmunity, and others | Varies | Varies |
| Vaccine-induced immune thrombotic thrombocytopenia (VITT) | Platelets, immune cells producing anti-PF4 antibodies | Formation of antibodies against platelet factor 4 (PF4), complement activation | Thrombosis, thrombocytopenia | Post-vaccination (variable) |
| Vaccine-associated enhanced disease (VAED) | B cells, Th2 skewed immunity | Non-neutralizing or suboptimal antibodies, generated in response to a vaccine, facilitate viral entry into host cells through Fc receptors or complement receptors | Worsening of diseases/symptoms | Upon exposure to natural virus after vaccination |
| Other | APCs, B cells, T cells | Activation of immune cells and release of cytokines (IL-1, IL-6, IL-12, TNF-α) | Varies | Post-vaccination |
NOTES: APCs: Antigen-Presenting Cells; IFN: Interferon, IL: interleukin, PF4: platelet factor 4, TNF: Tumor Necrosis Factor.
SOURCES: Chen et al., 2022b; Dabbiru et al., 2023; Lamprinou et al., 2023; Segal and Shoenfeld, 2018.
In contrast, delayed-type hypersensitivity reactions involve a different immune pathway. T cells, particularly CD4+ helper T cells, are central to these reactions. Upon exposure to an antigen that the immune system has seen before, T cells secrete cytokines, such as IFNγ, IL-2, and tumor necrosis factor-alpha (TNF-α). The symptoms associated with this reaction, such as rash, fever, and joint pain, typically develop days to weeks after vaccination, distinguishing them from the immediate-type reactions (Biedermann et al., 2000).
Autoimmune reactions in the context of vaccination encompass a variety of plausible mechanisms (Chen et al., 2022a; Lamprinou et al., 2023):
- Molecular mimicry, where vaccine antigens closely resemble the body’s own proteins, potentially leading to the production of autoantibodies or autoreactive T cells that target self-tissues (Segal and Shoenfeld, 2018).
- Bystander activation, when localized inflammation exposes self-antigens, leading to the activation of previously dormant self-reactive lymphocytes.
- Epitope spreading, particularly with repeat vaccinations, where the initial immune response to vaccine antigens broadens to include self-antigens.
- Polyclonal activation and adjuvant-induced autoimmunity, where intense immune stimulation, potentially exacerbated by adjuvants, overcomes the tolerance to self-antigens, resulting in autoimmunity.
A current and significant concern is vaccine-induced immune thrombotic thrombocytopenia (VITT), an extremely rare condition characterized by forming antibodies against platelet factor 4 (PF4). This activates platelets and immune cells producing anti-PF4 antibodies (Dabbiru et al., 2023). The role of complement activation in promoting a prothrombotic state is also being explored (see Chapter 5).
Vaccine-associated enhanced disease (VAED) and antibody-dependent enhancement (ADE) are critical considerations in vaccine development, particularly highlighted by historical challenges with the formalin-inactivated respiratory syncytial virus (RSV) vaccine (Acosta et al., 2015). VAED encompasses a spectrum of phenomena where vaccination paradoxically exacerbates the disease upon exposure to the natural pathogen, mediated through mechanisms such as ADE. In ADE, non-neutralizing or suboptimal antibodies generated by the vaccine facilitate the pathogen’s entry into host cells via Fc receptors, leading to increased viral replication and severe disease manifestations (Gartlan et al., 2022). The formalin-inactivated RSV vaccine is a notable example where immunization-induced antibodies not only failed to confer protection but also potentiated respiratory disease upon subsequent natural RSV infection. This outcome was partly attributed to the vaccine eliciting a skewed Th2-type immune response, promoting eosinophilic infiltration and severe lung pathology, rather than a protective Th1-type response (Gartlan et al., 2022). Additionally, immune complexes formed by the vaccine-induced antibodies could activate complement pathways, contributing to tissue damage.
Furthermore, general vaccine reactions encompass a wide array of immune responses. These involve the activation of antigen-presenting cells, B cells, and T cells. Cytokines, such as IL-1, IL-6, IL-12, and TNF-α, play a significant role in the initial immune response to vaccines, contributing to both their protective effects and potential harms.
ADJUVANTS
Adjuvants in vaccines serve to enhance the body’s immune response to an antigen, ensuring a stronger and longer-lasting immunity by activating TLRs on antigen-presenting cells to stimulate a strong innate immune response that produces a strong adaptive immune response. For example, aluminum salts create a depot effect for sustained antigen release, and oil-in-water emulsions, such as MF59, increase cytokine release and antigen uptake (Wilkins et al., 2017). Adjuvants such as AS01, AS02, AS03, and saponins stimulate antigen-presenting cells, such as dendritic cells, to activate T cells, and CpG oligodeoxynucleotides activate TLR9 (Facciola et al., 2022). These mechanisms, although crucial for vaccine efficacy, can sometimes lead to adverse reactions, primarily localized ones, such as inflammation and soreness, due to heightened immune activation at the site of injection. Table 2-3 lists some of the most commonly used adjuvants and their mechanisms of action.
TABLE 2-3 Most Commonly Used Adjuvants in Vaccines
| Adjuvant | Mechanism of Action |
|---|---|
| Aluminum salts (alum) | Creates a depot effect, slowly releasing antigen and enhancing antigen uptake by antigen-presenting cells. |
| MF59 (oil-in-water emulsion) | Increases cytokine release and antigen uptake, stimulating a stronger immune response. |
| AS04 (aluminum salt + monophosphoryl lipid [MPL] A) | Combines alum’s depot effect with MPL to enhance the immune response. MPL activates TLR4. |
| CpG oligodeoxynucleotides | Activates TLR9, enhancing the immune response to specific pathogens. |
| AS01 (liposome based) | Stimulates dendritic cells and T cell responses, enhancing both innate and adaptive immunity. |
| Virosomes | Mimics viral infection, enhancing the immune system’s recognition and response to the antigen. |
| QS-21 (saponin based) | Enhances antigen presentation and stimulates both humoral and cellular immune responses. |
| Lipopolysaccharide (LPS) | Activates TLR4, which leads to strong antibody responses by activating Th2 cells. |
| Poly-ICLC (polyinosinicpolycytidylic acid) | Mimics viral RNA, stimulating a strong immune response. |
| Matrix-M® (saponin based) | Activates antigen-presenting cells and boosts cytokine production, enhancing T cell and antibody responses. |
| Adjuvant system 03 (AS03, oil-in-water emulsion) | Contains squalene, DL-α-tocopherol, and polysorbate 80, enhancing immune response via cytokine modulation. |
SOURCES: Stertman et al., 2023; Wilkins et al., 2017.
Matrix-M® is a saponin-based adjuvant in NVX-CoV2373, which is the only specifically adjuvanted COVID-19 vaccine. It consists of Quillaja Saponaria Molina extracts, known for their ability to stimulate both the innate and adaptive arms of the immune system. It enhances immune responses by activating antigen-presenting cells and boosting cytokine production, which facilitates a stronger T cell and antibody response to the vaccine antigen (Stertman et al., 2023). Its mechanism of action increases the vaccine’s efficacy, but like other adjuvants, it can also contribute to or cause reactions. In the United States, FDA approves adjuvants only as components of vaccines, not as stand-alone products, because their properties can vary based on their concentration and interaction with other ingredients in the vaccine formulation.
Potential harms of vaccination necessitate a thorough investigation of mechanisms. Examining their immune response will help investigators gain insights into possible mechanisms of vaccine-related harms.
Through an examination of clinical trials, epidemiology studies, case reports, preclinical in vivo and in silico work, and insights from animal models, the committee has delved into possible mechanisms that may contribute to adverse events. Understanding these mechanisms is paramount in ensuring the safety and well-being of individuals receiving COVID-19 vaccines.
REFERENCES
Acosta, P. L., M. T. Caballero, and F. P. Polack. 2015. Brief history and characterization of enhanced respiratory syncytial virus disease. Clinical and Vaccine Immunology 23(3):189–195. https://doi.org/10.1128/cvi.00609-15.
Alameh, M. G., I. Tombacz, E. Bettini, K. Lederer, C. Sittplangkoon, J. R. Wilmore, B. T. Gaudette, O. Y. Soliman, M. Pine, P. Hicks, T. B. Manzoni, J. J. Knox, J. L. Johnson, D. Laczko, H. Muramatsu, B. Davis, W. Meng, A. M. Rosenfeld, S. Strohmeier, P. J. C. Lin, B. L. Mui, Y. K. Tam, K. Karikó, A. Jacquet, F. Krammer, P. Bates, M. P. Cancro, D. Weissman, E. T. Luning Prak, D. Allman, M. Locci, and N. Pardi. 2021. Lipid nanoparticles enhance the efficacy of mRNA and protein subunit vaccines by inducing robust T follicular helper cell and humoral responses. Immunity 54(12):2877–2892. https://doi.org/10.1016/j.immuni.2021.11.001.
Bettini, E., and M. Locci. 2021. SARS-CoV-2 mRNA vaccines: Immunological mechanism and beyond. Vaccines 9(2). https://doi.org/10.3390/vaccines9020147.
Biedermann, T., M. Kneilling, R. Mailhammer, K. Maier, C. A. Sander, G. Kollias, S. L. Kunkel, L. Hultner, and M. Rocken. 2000. Mast cells control neutrophil recruitment during T cell-mediated delayed-type hypersensitivity reactions through tumor necrosis factor and macrophage inflammatory protein 2. Journal of Experimental Medicine 192(10):1441–1452. https://doi.org/10.1084/jem.192.10.1441.
Bos, R., L. Rutten, J. E. M. van der Lubbe, M. J. G. Bakkers, G. Hardenberg, F. Wegmann, D. Zuijdgeest, A. H. de Wilde, A. Koornneef, A. Verwilligen, D. van Manen, T. Kwaks, R. Vogels, T. J. Dalebout, S. K. Myeni, M. Kikkert, E. J. Snijder, Z. Li, D. H. Barouch, J. Vellinga, J. P. M. Langedijk, R. C. Zahn, J. Custers, and H. Schuitemaker. 2020. Ad26 vector–based COVID-19 vaccine encoding a prefusion-stabilized SARS-CoV-2 spike immunogen induces potent humoral and cellular immune responses. NPJ Vaccines 5:91. https://doi.org/10.1038/s41541-020-00243-x.
Bowen, J. E., A. C. Walls, A. Joshi, K. R. Sprouse, C. Stewart, M. A. Tortorici, N. M. Franko, J. K. Logue, I. G. Mazzitelli, S. W. Tiles, K. Ahmed, A. Shariq, G. Snell, N. T. Iqbal, J. Geffner, A. Bandera, A. Gori, R. Grifantini, H. Y. Chu, W. C. Van Voorhis, D. Corti, and D. Veesler. 2021. SARS-CoV-2 spike conformation determines plasma neutralizing activity. bioRxiv. https://doi.org/10.1101/2021.12.19.473391.
Bulcha, J. T., Y. Wang, H. Ma, P. W. L. Tai, and G. Gao. 2021. Viral vector platforms within the gene therapy landscape. Signal Transduction and Targeted Therapy 6(1):53. https://doi.org/10.1038/s41392-021-00487-6.
Cao, Y., and G. F. Gao. 2021. mRNA vaccines: A matter of delivery. EClinicalMedicine 32:100746. https://doi.org/10.1016/j.eclinm.2021.100746.
Chaplin, D. D. 2010. Overview of the immune response. Journal of Allergy and Clinical Immunology 125(2 Suppl 2):S3–S23. https://doi.org/10.1016/j.jaci.2009.12.980.
Chavda, V. P., R. Bezbaruah, D. Valu, B. Patel, A. Kumar, S. Prasad, B. B. Kakoti, A. Kaushik, and M. Jesawadawala. 2023. Adenoviral vector–based vaccine platform for COVID-19: Current status. Vaccines 11(2). https://doi.org/10.3390/vaccines11020432.
Chen, D. P., Y. H. Wen, W. T. Lin, and F. P. Hsu. 2022a. Association between the side effect induced by COVID-19 vaccines and the immune regulatory gene polymorphism. Frontiers in Immunology 13:941497. https://doi.org/10.3389/fimmu.2022.941497.
Chen, Y., Z. Xu, P. Wang, X. M. Li, Z. W. Shuai, D. Q. Ye, and H. F. Pan. 2022b. New-onset autoimmune phenomena post-COVID-19 vaccination. Immunology 165(4):386–401. https://doi.org/10.1111/imm.13443.
Chu, L., K. Vrbicky, D. Montefiori, W. Huang, B. Nestorova, Y. Chang, A. Carfi, D. K. Edwards, J. Oestreicher, H. Legault, F. J. Dutko, B. Girard, R. Pajon, J. M. Miller, R. Das, B. Leav, and R. McPhee. 2022. Immune response to SARS-CoV-2 after a booster of mRNA-1273: An open-label Phase 2 trial. Nature Medicine 28(5):1042–1049. https://doi.org/10.1038/s41591-022-01739-w.
Corbett, K. S., B. Flynn, K. E. Foulds, J. R. Francica, S. Boyoglu-Barnum, A. P. Werner, B. Flach, S. O’Connell, K. W. Bock, M. Minai, B. M. Nagata, H. Andersen, D. R. Martinez, A. T. Noe, N. Douek, M. M. Donaldson, N. N. Nji, G. S. Alvarado, D. K. Edwards, D. R. Flebbe, E. Lamb, N. A. Doria-Rose, B. C. Lin, M. K. Louder, S. O’Dell, S. D. Schmidt, E. Phung, L. A. Chang, C. Yap, J. M. Todd, L. Pessaint, A. Van Ry, S. Browne, J. Greenhouse, T. Putman-Taylor, A. Strasbaugh, T. A. Campbell, A. Cook, A. Dodson, K. Steingrebe, W. Shi, Y. Zhang, O. M. Abiona, L. Wang, A. Pegu, E. S. Yang, K. Leung, T. Zhou, I. T. Teng, A. Widge, I. Gordon, L. Novik, R. A. Gillespie, R. J. Loomis, J. I. Moliva, G. Stewart-Jones, S. Himansu, W. P. Kong, M. C. Nason, K. M. Morabito, T. J. Ruckwardt, J. E. Ledgerwood, M. R. Gaudinski, P. D. Kwong, J. R. Mascola, A. Carfi, M. G. Lewis, R. S. Baric, A. McDermott, I. N. Moore, N. J. Sullivan, M. Roederer, R. A. Seder, and B. S. Graham. 2020. Evaluation of the mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. New England Journal of Medicine 383(16):1544–1555. https://doi.org/10.1056/NEJMoa2024671.
Coughlan, L. 2020. Factors which contribute to the immunogenicity of non-replicating adenoviral vectored vaccines. Frontiers in Immunology 11:909. https://doi.org/10.3389/fimmu.2020.00909.
Creech, C. B., S. C. Walker, and R. J. Samuels. 2021. SARS-CoV-2 vaccines. JAMA 325(13):1318–1320. https://doi.org/10.1001/jama.2021.3199.
Cullis, P. R., and M. J. Hope. 2017. Lipid nanoparticle systems for enabling gene therapies. Molecular Therapy 25(7):1467–1475. https://doi.org/10.1016/j.ymthe.2017.03.013.
Dabbiru, V. A. S., L. Muller, L. Schonborn, and A. Greinacher. 2023. Vaccine-induced immune thrombocytopenia and thrombosis (VITT)—insights from clinical cases, in vitro studies and murine models. Journal of Clinical Medicine 12(19). https://doi.org/10.3390/jcm12196126.
Diamond, M. S., and T. C. Pierson. 2020. The challenges of vaccine development against a new virus during a pandemic. Cell Host & Microbe 27(5):699–703. https://doi.org/10.1016/j.chom.2020.04.021.
Ewer, K., S. Sebastian, A. J. Spencer, S. Gilbert, A. V. S. Hill, and T. Lambe. 2017. Chimpanzee adenoviral vectors as vaccines for outbreak pathogens. Human Vaccines & Immunotherapeutics 13(12):3020–3032. https://doi.org/10.1080/21645515.2017.1383575.
Facciola, A., G. Visalli, A. Lagana, and A. Di Pietro. 2022. An overview of vaccine adjuvants: Current evidence and future perspectives. Vaccines 10(5). https://doi.org/10.3390/vaccines10050819.
FDA (Food and Drug Administration). 2021. BLA clinical review memorandum—COMIRNATY. https://www.fda.gov/media/152256/download (accessed December 12, 2023).
FDA. 2022. BLA clinical review memorandum—SPIKEVAX. https://www.fda.gov/media/156342/download (accessed December 12, 2023).
Fitzgerald, K. A., and J. C. Kagan. 2020. Toll-like receptors and the control of immunity. Cell 180(6):1044–1066. https://doi.org/10.1016/j.cell.2020.02.041.
Gartlan, C., T. Tipton, F. J. Salguero, Q. Sattentau, A. Gorringe, and M. W. Carroll. 2022. Vaccine-associated enhanced disease and pathogenic human coronaviruses. Frontiers in Immunology 13:882972. https://doi.org/10.3389/fimmu.2022.882972.
Geisbert, T. W., M. Bailey, L. Hensley, C. Asiedu, J. Geisbert, D. Stanley, A. Honko, J. Johnson, S. Mulangu, M. G. Pau, J. Custers, J. Vellinga, J. Hendriks, P. Jahrling, M. Roederer, J. Goudsmit, R. Koup, and N. J. Sullivan. 2011. Recombinant adenovirus serotype 26 (Ad26) and Ad35 vaccine vectors bypass immunity to ad5 and protect nonhuman primates against ebolavirus challenge. Journal of Virology 85(9):4222–4233. https://doi.org/10.1128/jvi.02407-10.
Jackson, L. A., E. J. Anderson, N. G. Rouphael, P. C. Roberts, M. Makhene, R. N. Coler, M. P. McCullough, J. D. Chappell, M. R. Denison, L. J. Stevens, A. J. Pruijssers, A. McDermott, B. Flach, N. A. Doria-Rose, K. S. Corbett, K. M. Morabito, S. O’Dell, S. D. Schmidt, P. A. Swanson, II, M. Padilla, J. R. Mascola, K. M. Neuzil, H. Bennett, W. Sun, E. Peters, M. Makowski, J. Albert, K. Cross, W. Buchanan, R. Pikaart-Tautges, J. E. Ledgerwood, B. S. Graham, J. H. Beigel, and mRNA-1273 Study Group. 2020. An mRNA vaccine against SARS-CoV-2—preliminary report. New England Journal of Medicine 383(20):1920–1931. https://doi.org/10.1056/NEJMoa2022483.
Jarvis, D. L. 2003. Developing baculovirus-insect cell expression systems for humanized recombinant glycoprotein production. Virology 310(1):1–7. https://doi.org/10.1016/s0042-6822(03)00120-x.
Karikó, K., H. Muramatsu, F. A. Welsh, J. Ludwig, H. Kato, S. Akira, and D. Weissman. 2008. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Molecular Therapy 16(11):1833–1840. https://doi.org/10.1038/mt.2008.200.
Keech, C., G. Albert, I. Cho, A. Robertson, P. Reed, S. Neal, J. S. Plested, M. Zhu, S. Cloney-Clark, H. Zhou, G. Smith, N. Patel, M. B. Frieman, R. E. Haupt, J. Logue, M. McGrath, S. Weston, P. A. Piedra, C. Desai, K. Callahan, M. Lewis, P. Price-Abbott, N. Formica, V. Shinde, L. Fries, J. D. Lickliter, P. Griffin, B. Wilkinson, and G. M. Glenn. 2020. Phase 1–2 trial of a SARS-CoV-2 recombinant spike protein nanoparticle vaccine. New England Journal of Medicine 383(24):2320–2332. https://doi.org/10.1056/NEJMoa2026920.
Khehra, N., I. Padda, U. Jaferi, H. Atwal, S. Narain, and M. S. Parmar. 2021. Tozinameran (BNT162b2) vaccine: The journey from preclinical research to clinical trials and authorization. AAPS PharmSciTech 22(5):172. https://doi.org/10.1208/s12249-021-02058-y.
Kim, S. C., S. S. Sekhon, W. R. Shin, G. Ahn, B. K. Cho, J. Y. Ahn, and Y. H. Kim. 2022. Modifications of mRNA vaccine structural elements for improving mRNA stability and translation efficiency. Molecular & Cellular Toxicology 18(1):1–8. https://doi.org/10.1007/s13273-021-00171-4.
Kirchdoerfer, R. N., C. A. Cottrell, N. Wang, J. Pallesen, H. M. Yassine, H. L. Turner, K. S. Corbett, B. S. Graham, J. S. McLellan, and A. B. Ward. 2016. Pre-fusion structure of a human coronavirus spike protein. Nature 531(7592):118–121. https://doi.org/10.1038/nature17200.
Krammer, F. 2020. SARS-CoV-2 vaccines in development. Nature 586(7830):516–527. https://doi.org/10.1038/s41586-020-2798-3.
Laczko, D., M. J. Hogan, S. A. Toulmin, P. Hicks, K. Lederer, B. T. Gaudette, D. Castano, F. Amanat, H. Muramatsu, T. H. Oguin, III, A. Ojha, L. Zhang, Z. Mu, R. Parks, T. B. Manzoni, B. Roper, S. Strohmeier, I. Tombacz, L. Arwood, R. Nachbagauer, K. Karikó, J. Greenhouse, L. Pessaint, M. Porto, T. Putman-Taylor, A. Strasbaugh, T. A. Campbell, P. J. C. Lin, Y. K. Tam, G. D. Sempowski, M. Farzan, H. Choe, K. O. Saunders, B. F. Haynes, H. Andersen, L. C. Eisenlohr, D. Weissman, F. Krammer, P. Bates, D. Allman, M. Locci, and N. Pardi. 2020. A single immunization with nucleoside-modified mRNA vaccines elicits strong cellular and humoral immune responses against SARS-CoV-2 in mice. Immunity 53(4):724–732. https://doi.org/10.1016/j.immuni.2020.07.019.
Laidlaw, B. J., and A. H. Ellebedy. 2022. The germinal centre B cell response to SARS-CoV-2. Nature Reviews: Immunology 22(1):7–18. https://doi.org/10.1038/s41577-021-00657-1.
Lamprinou, M., A. Sachinidis, E. Stamoula, T. Vavilis, and G. Papazisis. 2023. COVID-19 vaccines adverse events: Potential molecular mechanisms. Immunologic Research 71(3):356–372. https://doi.org/10.1007/s12026-023-09357-5.
Lederer, K., D. Castano, D. Gomez Atria, T. H. Oguin, III, S. Wang, T. B. Manzoni, H. Muramatsu, M. J. Hogan, F. Amanat, P. Cherubin, K. A. Lundgreen, Y. K. Tam, S. H. Y. Fan, L. C. Eisenlohr, I. Maillard, D. Weissman, P. Bates, F. Krammer, G. D. Sempowski, N. Pardi, and M. Locci. 2020. SARS-CoV-2 mRNA vaccines foster potent antigen-specific germinal center responses associated with neutralizing antibody generation. Immunity 53(6):1281–1295. https://doi.org/10.1016/j.immuni.2020.11.009.
Lukashev, A. N., and A. A. Zamyatnin, Jr. 2016. Viral vectors for gene therapy: Current state and clinical perspectives. Biochemistry 81(7):700–708. https://doi.org/10.1134/S0006297916070063.
Maier, M. A., M. Jayaraman, S. Matsuda, J. Liu, S. Barros, W. Querbes, Y. K. Tam, S. M. Ansell, V. Kumar, J. Qin, X. Zhang, Q. Wang, S. Panesar, R. Hutabarat, M. Carioto, J. Hettinger, P. Kandasamy, D. Butler, K. G. Rajeev, B. Pang, K. Charisse, K. Fitzgerald, B. L. Mui, X. Du, P. Cullis, T. D. Madden, M. J. Hope, M. Manoharan, and A. Akinc. 2013. Biodegradable lipids enabling rapidly eliminated lipid nanoparticles for systemic delivery of RNAI therapeutics. Molecular Therapy 21(8):1570–1578. https://doi.org/10.1038/mt.2013.124.
Marfe, G., S. Perna, and A. K. Shukla. 2021. Effectiveness of COVID-19 vaccines and their challenges (review). Experimental and Therapeutic Medicine 22(6):1407. https://doi.org/10.3892/etm.2021.10843.
Martinez-Flores, D., J. Zepeda-Cervantes, A. Cruz-Resendiz, S. Aguirre-Sampieri, A. Sampieri, and L. Vaca. 2021. SARSCoV-2 vaccines based on the spike glycoprotein and implications of new viral variants. Frontiers in Immunology 12:701501. https://doi.org/10.3389/fimmu.2021.701501.
McLeod, J. J., B. Baker, and J. J. Ryan. 2015. Mast cell production and response to IL-4 and IL-13. Cytokine 75(1):57–61. https://doi.org/10.1016/j.cyto.2015.05.019.
Menegale, F., M. Manica, A. Zardini, G. Guzzetta, V. Marziano, V. d’Andrea, F. Trentini, M. Ajelli, P. Poletti, and S. Merler. 2023. Evaluation of waning of SARS-CoV-2 vaccine-induced immunity: A systematic review and meta-analysis. JAMA Network Open 6(5):e2310650. https://doi.org/10.1001/jamanetworkopen.2023.10650.
Ndeupen, S., Z. Qin, S. Jacobsen, A. Bouteau, H. Estanbouli, and B. Z. Igyarto. 2021. The mRNA-LNP platform’s lipid nanoparticle component used in preclinical vaccine studies is highly inflammatory. iScience 24(12):103479. https://doi.org/10.1016/j.isci.2021.103479.
Pallesen, J., N. Wang, K. S. Corbett, D. Wrapp, R. N. Kirchdoerfer, H. L. Turner, C. A. Cottrell, M. M. Becker, L. Wang, W. Shi, W. P. Kong, E. L. Andres, A. N. Kettenbach, M. R. Denison, J. D. Chappell, B. S. Graham, A. B. Ward, and J. S. McLellan. 2017. Immunogenicity and structures of a rationally designed prefusion MERS-COV spike antigen. Proceedings of the National Academy of Sciences of the United States of America 114(35):E7348–E7357. https://doi.org/10.1073/pnas.1707304114.
Pardi, N., S. Tuyishime, H. Muramatsu, K. Karikó, B. L. Mui, Y. K. Tam, T. D. Madden, M. J. Hope, and D. Weissman. 2015. Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes. Journal of Controlled Release 217:345–351. https://doi.org/10.1016/j.jconrel.2015.08.007.
Pardi, N., M. J. Hogan, F. W. Porter, and D. Weissman. 2018. mRNA vaccines—a new era in vaccinology. Nature Reviews Drug Discovery 17(4):261–279. https://doi.org/10.1038/nrd.2017.243.
Pettini, E., D. Medaglini, and A. Ciabattini. 2022. Profiling the B cell immune response elicited by vaccination against the respiratory virus SARS-CoV-2. Frontiers in Immunology 13:1058748. https://doi.org/10.3389/fimmu.2022.1058748.
Pollard, A. J., and E. M. Bijker. 2021. A guide to vaccinology: From basic principles to new developments. Nature Reviews: Immunology 21(2):83–100. https://doi.org/10.1038/s41577-020-00479-7.
Robbiani, D. F., C. Gaebler, F. Muecksch, J. C. C. Lorenzi, Z. Wang, A. Cho, M. Agudelo, C. O. Barnes, A. Gazumyan, S. Finkin, T. Hagglof, T. Y. Oliveira, C. Viant, A. Hurley, H. H. Hoffmann, K. G. Millard, R. G. Kost, M. Cipolla, K. Gordon, F. Bianchini, S. T. Chen, V. Ramos, R. Patel, J. Dizon, I. Shimeliovich, P. Mendoza, H. Hartweger, L. Nogueira, M. Pack, J. Horowitz, F. Schmidt, Y. Weisblum, E. Michailidis, A. W. Ashbrook, E. Waltari, J. E. Pak, K. E. Huey-Tubman, N. Koranda, P. R. Hoffman, A. P. West, Jr., C. M. Rice, T. Hatziioannou, P. J. Bjorkman, P. D. Bieniasz, M. Caskey, and M. C. Nussenzweig. 2020. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature 584(7821):437–442. https://doi.org/10.1038/s41586-020-2456-9.
Sadarangani, M., A. Marchant, and T. R. Kollmann. 2021. Immunological mechanisms of vaccine-induced protection against COVID-19 in humans. Nature Reviews: Immunology 21(8):475–484. https://doi.org/10.1038/s41577-021-00578-z.
Sadoff, J., G. Gray, A. Vandebosch, V. Cárdenas, G. Shukarev, B. Grinsztejn, P. A. Goepfert, C. Truyers, H. Fennema, B. Spiessens, K. Offergeld, G. Scheper, K. L. Taylor, M. L. Robb, J. Treanor, D. H. Barouch, J. Stoddard, M. F. Ryser, M. A. Marovich, K. M. Neuzil, L. Corey, N. Cauwenberghs, T. Tanner, K. Hardt, J. Ruiz-Guiñazú, M. Le Gars, H. Schuitemaker, J. Van Hoof, F. Struyf, and M. Douoguih. 2021. Safety and efficacy of single-dose Ad26.COV2.S vaccine against COVID-19. New England Journal of Medicine 384(23):2187–2201. https://doi.org/10.1056/NEJMoa2101544.
Segal, Y., and Y. Shoenfeld. 2018. Vaccine-induced autoimmunity: The role of molecular mimicry and immune crossreaction. Cellular & Molecular Immunology 15(6):586–594. https://doi.org/10.1038/cmi.2017.151.
Stertman, L., A. E. Palm, B. Zarnegar, B. Carow, C. Lunderius Andersson, S. E. Magnusson, C. Carnrot, V. Shinde, G. Smith, G. Glenn, L. Fries, and K. Lovgren Bengtsson. 2023. The matrix-M adjuvant: A critical component of vaccines for the 21st century. Human Vaccines & Immunotherapeutics 19(1):2189885. https://doi.org/10.1080/21645515.2023.2189885.
Stone, C. A., Jr., C. R. F. Rukasin, T. M. Beachkofsky, and E. J. Phillips. 2019. Immune-mediated adverse reactions to vaccines. British Journal of Clinical Pharmacology 85(12):2694–2706. https://doi.org/10.1111/bcp.14112.
Tam, H. H., M. B. Melo, M. Kang, J. M. Pelet, V. M. Ruda, M. H. Foley, J. K. Hu, S. Kumari, J. Crampton, A. D. Baldeon, R. W. Sanders, J. P. Moore, S. Crotty, R. Langer, D. G. Anderson, A. K. Chakraborty, and D. J. Irvine. 2016. Sustained antigen availability during germinal center initiation enhances antibody responses to vaccination. Proceedings of the National Academy of Sciences of the United States of America 113(43):E6639–E6648. https://doi.org/10.1073/pnas.1606050113.
Teijaro, J. R., and D. L. Farber. 2021. COVID-19 vaccines: Modes of immune activation and future challenges. Nature Reviews: Immunology 21(4):195–197. https://doi.org/10.1038/s41577-021-00526-x.
Trougakos, I. P., E. Terpos, H. Alexopoulos, M. Politou, D. Paraskevis, A. Scorilas, E. Kastritis, E. Andreakos, and M. A. Dimopoulos. 2022. Adverse effects of COVID-19 mRNA vaccines: The spike hypothesis. Trends in Molecular Medicine 28(7):542–554. https://doi.org/10.1016/j.molmed.2022.04.007.
Turner, J. S., J. A. O’Halloran, E. Kalaidina, W. Kim, A. J. Schmitz, J. Q. Zhou, T. Lei, M. Thapa, R. E. Chen, J. B. Case, F. Amanat, A. M. Rauseo, A. Haile, X. Xie, M. K. Klebert, T. Suessen, W. D. Middleton, P. Y. Shi, F. Krammer, S. A. Teefey, M. S. Diamond, R. M. Presti, and A. H. Ellebedy. 2021. SARS-CoV-2 mRNA vaccines induce persistent human germinal centre responses. Nature 596(7870):109–113. https://doi.org/10.1038/s41586-021-03738-2.
Verbeke, R., M. J. Hogan, K. Lore, and N. Pardi. 2022. Innate immune mechanisms of mRNA vaccines. Immunity 55(11):1993–2005. https://doi.org/10.1016/j.immuni.2022.10.014.
Walls, A. C., Y. J. Park, M. A. Tortorici, A. Wall, A. T. McGuire, and D. Veesler. 2020. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 183(6):1735. https://doi.org/10.1016/j.cell.2020.11.032.
Walsh, E. E., R. W. Frenck, Jr., A. R. Falsey, N. Kitchin, J. Absalon, A. Gurtman, S. Lockhart, K. Neuzil, M. J. Mulligan, R. Bailey, K. A. Swanson, P. Li, K. Koury, W. Kalina, D. Cooper, C. Fontes-Garfias, P. Y. Shi, O. Tureci, K. R. Tompkins, K. E. Lyke, V. Raabe, P. R. Dormitzer, K. U. Jansen, U. Sahin, and W. C. Gruber. 2020. Safety and immunogenicity of two RNA-based COVID-19 vaccine candidates. New England Journal of Medicine 383(25):2439–2450. https://doi.org/10.1056/NEJMoa2027906.
Watanabe, Y., L. Mendonca, E. R. Allen, A. Howe, M. Lee, J. D. Allen, H. Chawla, D. Pulido, F. Donnellan, H. Davies, M. Ulaszewska, S. Belij-Rammerstorfer, S. Morris, A. S. Krebs, W. Dejnirattisai, J. Mongkolsapaya, P. Supasa, G. R. Screaton, C. M. Green, T. Lambe, P. Zhang, S. C. Gilbert, and M. Crispin. 2021. Native-like SARS-CoV-2 spike glycoprotein expressed by ChAdOx1 nCoV-19/AZD1222 vaccine. ACS Central Science 7(4):594–602. https://doi.org/10.1021/acscentsci.1c00080.
Welsh, J. 2021. Coronavirus variants—will new mRNA vaccines meet the challenge? Engineering 7(6):712–714. https://doi.org/10.1016/j.eng.2021.04.005.
Widge, A. T., N. G. Rouphael, L. A. Jackson, E. J. Anderson, P. C. Roberts, M. Makhene, J. D. Chappell, M. R. Denison, L. J. Stevens, A. J. Pruijssers, A. B. McDermott, B. Flach, B. C. Lin, N. A. Doria-Rose, S. O’Dell, S. D. Schmidt, K. M. Neuzil, H. Bennett, B. Leav, M. Makowski, J. Albert, K. Cross, V. V. Edara, K. Floyd, M. S. Suthar, W. Buchanan, C. J. Luke, J. E. Ledgerwood, J. R. Mascola, B. S. Graham, J. H. Beigel, and mRNA-1273 Study Group. 2021. Durability of responses after SARS-CoV-2 mRNA-1273 vaccination. New England Journal of Medicine 384(1):80–82. https://doi.org/10.1056/NEJMc2032195.
Wilkins, A. L., D. Kazmin, G. Napolitani, E. A. Clutterbuck, B. Pulendran, C. A. Siegrist, and A. J. Pollard. 2017. AS03- and Mf59adjuvanted influenza vaccines in children. Frontiers in Immunology 8:1760. https://doi.org/10.3389/fimmu.2017.01760.
Wrapp, D., N. Wang, K. S. Corbett, J. A. Goldsmith, C. L. Hsieh, O. Abiona, B. S. Graham, and J. S. McLellan. 2020. Cryo-em structure of the 2019-nCoV spike in the prefusion conformation. bioRxiv. https://doi.org/10.1101/2020.02.11.944462.
Xiaojie, S., L. Yu, Y. Lei, Y. Guang, and Q. Min. 2020. Neutralizing antibodies targeting SARS-CoV-2 spike protein. Stem Cell Research 50:102125. https://doi.org/10.1016/j.scr.2020.102125.
Yang, G., J. Wang, P. Sun, J. Qin, X. Yang, D. Chen, Y. Zhang, N. Zhong, and Z. Wang. 2023. SARS-CoV-2 epitope-specific T cells: Immunity response feature, TCR repertoire characteristics and cross-reactivity. Frontiers in Immunology 14:1146196. https://doi.org/10.3389/fimmu.2023.1146196.
Zost, S. J., P. Gilchuk, R. E. Chen, J. B. Case, J. X. Reidy, A. Trivette, R. S. Nargi, R. E. Sutton, N. Suryadevara, E. C. Chen, E. Binshtein, S. Shrihari, M. Ostrowski, H. Y. Chu, J. E. Didier, K. W. MacRenaris, T. Jones, S. Day, L. Myers, F. Eun-Hyung Lee, D. C. Nguyen, I. Sanz, D. R. Martinez, P. W. Rothlauf, L. M. Bloyet, S. P. J. Whelan, R. S. Baric, L. B. Thackray, M. S. Diamond, R. H. Carnahan, and J. E. Crowe, Jr. 2020. Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS-CoV-2 spike protein. Nature Medicine 26(9):1422–1427. https://doi.org/10.1038/s41591-020-0998-x.















