Chemical Ecology: The Chemistry of Biotic Interaction (1995)
Chapter: Supplementary Images
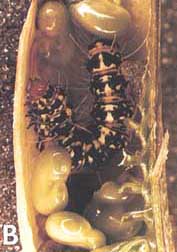
FIGURE 1 Arthropod chemical defenses.
A

(A) Blister beeble (Lytta polita) reflex-bleeding from the knee joints; the blood contains cantharidin, one of the first insect toxins characterized (4).
B

(B) Chrysomelid beetle larva (Plagiodera versicolora) emitting droplets of defensive secretion from its segmental glands; the fluid contains methylcyclopentanoid terpenes (chrysomelidial, plagiolactone) (5).
C

(C) Whip scorpion (Mastigoproctus giganteus) discharging an aimed jet of spray in response to pinching of an appendage with forceps; the spray contains 84% acetic acid (6).
D

(D) Millipede (Narceus gordanus) discharging its benzoquinone-containing defensive secretion from segmental glands (7).
E

(E) Ctenuchid moth (Cisseps Fulvicollis) discharging defensive droplets (chemistry unknown) from the cervical region.
F
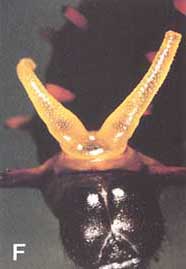
(F) Caterpillar of swallowtail butter fly (ß-selinene; selin-11-en-4a-ol) (8).
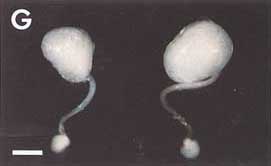
G

(G) Stalked eggs of green lacewing (Nodita floridona), showing droplets of defensive fluid (chemistry unknown) on stalks.
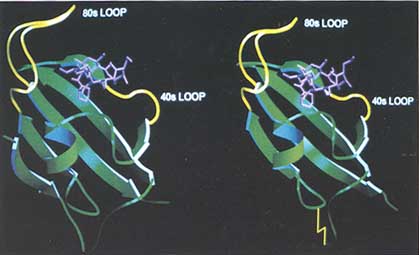
FIGURE 2 (Left) Diagram of the FKBP12-FK506 complex. The protein is shown by the ribbon convention and FK506 is shown as a ball-and-stick model. The ß-strand numbering referred to in the text is from bottom to top. (Right) Diagram of the FKBP13-FK506 complex. The disulfide bridge is shown as a yellow zigzag line. The N terminus of the protein is disordered and is not shown.

FIGURE 3 Crystal structure of the yeast transcriptional activator GCN4, bound to DNA (83). This structure confirms the prediction that had been based on the alignment of 11 sequences from a range of species that the conserved leucines (highlighted in yellow) would be oriented toward each other along the faces of neighboring helices. As predicted, the helices cross the DNA in the major groove. Surprisingly, the GCN4 residues making specific DNA base contacts (highlighted in red) are conserved among proteins with different DNA binding specificities, whereas the amino acids contacting the DNA backbone in the GCN4 structure (highlighted in blue) are less conserved in this protein family. Thus, while predictions made based upon linear amino acid sequences from 11 species were confirmed, we await three-dimensional structures from diverse sources to elucidate how variant amino acids alter the orientation of the conserved amino acids to effect base-sequence-specific recognition (figure by Tom Ellenberger).