Incorporating Integrated Diagnostics into Precision Oncology Care: Proceedings of a Workshop (2024)
Chapter: Proceedings of a Workshop
Proceedings of a Workshop
WORKSHOP OVERVIEW1
Advances in informatics and the diagnostic medical specialties (radiology, pathology, and laboratory medicine) could potentially reshape cancer diagnosis and enhance precision oncology care. Integrating multiple types of diagnostic data, combined with analysis with artificial intelligence (AI) algorithms, could help to guide personalized treatment and improve patient outcomes. No uniform strategy currently exists, however, to develop, validate, implement, and use integrated diagnostics in cancer care.
The National Cancer Policy Forum, in collaboration with the Computer Science and Telecommunications Board and the Board on Human–Systems Integration of the National Academies of Sciences, Engineering, and Medicine, hosted a public workshop on incorporating integrated diagnostics into precision oncology care on March 6 and 7, 2023. Hedvig Hricak, the Carroll and Milton Petrie endowed chair in the Department of Radiology at the Memorial Sloan Kettering Cancer Center (MSK), described the workshop as “an opportunity for the cancer community to discuss the current state of the
___________________
1 This workshop was organized by an independent planning committee whose role was limited to identification of topics and speakers. This Proceedings of a Workshop was prepared by the rapporteurs as a factual summary of the presentations and discussions that took place at the workshop. Statements, recommendations, and opinions expressed are those of individual presenters and participants and are not endorsed or verified by the National Academies of Sciences, Engineering, and Medicine, and they should not be construed as reflecting any group consensus.
field of integrated diagnostics, including the purpose, goals, and components of integrated diagnostics.” It featured presentations and panel discussions on a range of topics, including:
- current efforts to develop and implement integrated diagnostics to inform treatment decision making in cancer care;
- implications of integrated diagnostics for clinician training, care workflows, and the organization of care teams;
- opportunities for evidence generation to inform validation, clinical utility, regulatory oversight, and insurance coverage for integrated diagnostics;
- strategies for ongoing quality assurance, evaluation, and refinement of integrated cancer diagnostics based on new evidence; and
- mechanisms to enable broad patient access to integrated diagnostics, particularly in community-based settings of cancer care.
This Proceedings of a Workshop summarizes the presentations and discussions from the workshop. Observations and suggestions from individual participants are discussed throughout the proceedings and highlighted in Boxes 1 and 2. Appendixes A and B provide the Statement of Task and agenda, respectively. Speaker presentations and the workshop webcast are archived online.2
OVERVIEW OF THE CURRENT STATUS OF AND VISION FOR INTEGRATED DIAGNOSTICS
An Evolving Integrated Data Science Approach to Personalized Oncology Care
There is currently no consensus definition of integrated diagnostics, said Kojo Elenitoba-Johnson, inaugural chair of the department of pathology and laboratory medicine and James Ewing Alumni Chair of Pathology at MSK. He referred to Hricak’s working definition as “the convergence of imaging, pathology, and laboratory testing, supplemented by advanced information technology, which has enormous potential for revolutionizing the diagnosis and therapeutic management of many diseases, including cancer.”
___________________
2 See https://www.nationalacademies.org/event/03-06-2023/incorporating-integrated-diagnostics-into-precision-oncology-care-a-workshop (accessed May 26, 2023).
BOX 1
Observations on the Development and Use of Integrated Diagnostics in Cancer Care Made by Individual Workshop Participants
Describing Integrated Diagnostics
- The concept of integrated diagnostics refers to the convergence of imaging, pathology, and laboratory testing data, augmented with information technology. (Elenitoba-Johnson, Hricak)
- Integrated diagnostics are not a clinical decision support system (although they support clinical decision making), and they are much more than a data aggregation dashboard. (Hricak)
- Integrated diagnostics are intended to be patient centered. (Elenitoba-Johnson, Hirsch, Hricak, Lennerz, Osterman)
Diagnostics Research and Practice
- Diagnostic testing is siloed by discipline, making it challenging for oncology clinicians to assimilate patient data to inform a patient’s care plan. (Hricak)
- Multidisciplinary teams are essential to develop, implement, and use integrated diagnostics. (Hricak, Osterman, Shah, Thai)
- The goal of integrated diagnostics is to create clinical partnerships and align workflows to facilitate the integration of diagnostic data from across disciplines. (Lennerz, Salto-Tellez, Schnall)
- Cultural barriers to implementing integrated diagnostics persist; despite growing evidence supporting the use of artificial intelligence (AI) in precision oncology, many clinicians still hesitate to do so in practice. (Choudhury, Elenitoba-Johnson, Krestin, Oyer)
- Key factors that affect clinician adoption of AI-based integrated diagnostics are trust in the technology and accountability. (Choudhury, Dorr)
- Incentives are needed to promote the adoption of integrated diagnostics into practice. (Elenitoba-Johnson)
Evidence Generation
- Developing a learning health care system is one option to generate the evidence needed to evaluate integrated diagnostics. (Levy)
- Electronic health record (EHR) data and capabilities can be leveraged to support the evaluation of integrated diagnostics. (Levy, Meropol)
-
- EHR data collected during routine care can be used for pragmatic studies. (Sawyers)
- Intentional EHR-based data collection can support prospective observational studies (i.e., data that may not be needed for a patient’s routine care but are collected as part of routine workflows). (Meropol, Sawyers)
Design and Use in Clinical Practice
AI Models
- AI models in diagnostics are becoming increasingly computationally complex but also less transparent. (Thai)
- AI algorithms can perpetuate and amplify biases in training datasets, leading to biased interpretations when applied to broader populations. (Dorr, Thai)
- An AI algorithm developed at one institution often does not perform as expected or required at different institutions. (Barzilay, Levy, Salto-Tellez)
- Machine learning algorithms that are incorporated into clinical workflow can nudge clinicians toward testing or treatment options based on patient results, new research, or other information. (Robison)
- Self-supervised learning by AI is one approach to handling the massive volumes of health data being generated by medical technologies and devices. (Comaniciu)
Workflow and Workforce
- The use of unstructured, text-based clinical notes in the EHR persists, and the lack of uniform, structured, and synoptic reporting contributes to diagnostic uncertainty. (Hricak, Osterman)
- Data standards exist, but entry of structured data does not always fit well within clinical workflow. (Osterman)
- Patient-centric user interfaces for integrated diagnostics are needed, using design principles to prioritize the most important information for patient care and actions to consider. (Dorr, Robison, Shah)
- A diagnostic management team approach can improve care and reduce medical errors by providing guidance on appropriate diagnostic test ordering and use and interpretation of test results. (Laposata)
- Investments in data engineering are needed to scale integrated diagnostics. (Shah)
Regulatory and Coverage Considerations
- The U.S. Food and Drug Administration evaluates integrated diagnostics and AI-based digital technologies according to the traditional risk-based review framework for medical devices. (Philip)
- Clinical coverage for a service generally requires a high level of evidence of clinical utility, with clearly defined improvement in outcome (which might be an intermediate outcome for diagnostics). (Malin)
- Billing codes are now available for the time laboratory medicine experts spend advising clinicians on diagnostic test selection and result interpretation. (Laposata)
Access and Equity
- Equity is central to all efforts in developing and using integrated diagnostics. (Nilsen)
- Validating AI-based risk prediction algorithms across institutions and populations is a challenge. (Barzilay)
- One algorithmic approach to improve equity in AI model design is distributional shift detection, in which the algorithm learns to detect bias automatically. (Barzilay)
- Because it may not be possible to explain how an algorithm makes a prediction, an alternative approach is to design the model to learn and report when its prediction cannot be trusted, based on calibrated uncertainty. (Barzilay)
- Most patients with cancer receive their care in community settings, and disparities in access to high-quality care and new technologies persist in many communities outside the catchment area of National Cancer Institute–designated cancer centers or other major academic centers. (Brawley, Oyer, Shulman)
- Introducing new technologies into clinical practice can create disparities in care (e.g., due to lack of access, uneven implementation, and the use of limited resources for unnecessary care). (Brawley)
NOTE: This list is the rapporteurs’ synopsis of observations made by one or more individual speakers as identified. These statements have not been endorsed or verified by the National Academies of Sciences, Engineering, and Medicine. They are not intended to reflect a consensus among workshop participants.
BOX 2
Opportunities to Advance the Development, Implementation, and Use of Integrated Diagnostics in Precision Cancer Care Suggested by Individual Workshop Participants
Implementing Integrated Diagnostics
- Ensure interoperability of data systems across clinical disciplines. (Osterman, Trentadue)
- Establish, adopt, and integrate data standards into clinical workflows. (Osterman)
- Deploy structured electronic health record fields for standardized data elements. (Osterman)
- Adopt synoptic operative reports. (Hricak, Osterman)
- Develop a lexicon to convey degree of diagnostic certainty. (Hricak)
- Incorporate patient-level longitudinal data. (Levy, Robison)
- Collaborate to advance the process in automating tumor imaging annotation and segmentation. (Hricak)
- Develop institutional practices for data governance and facilitating a culture of data sharing. (Butte, Hricak)
- Ensure integrated diagnostics have feedback loops to support a learning health care system. (Kronander, Levy)
- Recruit more data scientists to cancer research and care to enhance efficient translation of scientific and technological advances in patient care. (Shah)
Improving Design and Use
- Use training and testing datasets that are representative of the population for which the technology is intended. (Barzilay Levy, Thai)
- Develop a code of conduct for implementing AI-based integrated diagnostics. (Dorr)
- Establish shared or distributed accountability based on risk and intended use. (Choudhury)
- Promote open data policies, requiring public release of data used in algorithm training and testing. (Shah)
- Use principles of risk communication to support shared decision making (e.g., use numbers; avoid imprecise terms; use data visualizations to convey risk; keep denominators consistent; discuss absolute and not relative risk). (Politi)
- Apply implementation science principles to promote the uptake of integrated diagnostics into clinical workflows and prevent unintended consequences. (Dorr)
- Train the next generation of scientists and clinicians to be able to communicate across disciplines and apply an integrated approach to diagnostics. (Gold, Schnall)
- Embrace regulatory science, and actively engage with regulators on issues related to the evaluation and implementation of integrated diagnostics. (Lennerz)
- Provide insurance reimbursement for patient diagnosis and care rather than conducting specific diagnostic tests and procedures. (Kronander)
Facilitating Access and Equity
- Build a national hub-and-spoke system to address gaps in access to high-quality cancer care to facilitate the equitable deployment of integrated diagnostics. (Oyer)
- Apply implementation science approaches to promote equitable uptake and diffusion of new tools and technologies and avoid worsening disparities in resource-poor settings. (Brawley)
- Leverage new digital technologies to increase the capacity and effectiveness of the cancer care workforce and improve patient experience and outcomes. (Oyer)
- Provide training to increase the ability and skills of the current oncology workforce to integrate diagnostic tools into practice. (Oyer)
NOTE: This list is the rapporteurs’ synopsis of suggestions made by one or more individual speakers as identified. These statements have not been endorsed or verified by the National Academies of Sciences, Engineering, and Medicine. They are not intended to reflect a consensus among workshop participants.
Elenitoba-Johnson said the core patient-centered goals for integrated diagnostics include:
- Diagnosis: “What disease does the patient have?”
- Residual disease: “How much disease is there?”
- Prognosis: “Who needs treatment?”
- Therapy: “What is the best treatment for this patient?”
- Pharmacodynamics/pharmacogenomics: “What dose?”
He characterized the overall goal of integrated diagnostics as “precision diagnostics scaled at a population level for impact at an individual level.”
Integrated diagnostics present a range of opportunities to reduce medical errors and improve patient outcomes, noted Hricak. From a clinical perspective, she said integrated diagnostics can function as AI-facilitated tumor boards.3 For research, large annotated and curated databases can provide opportunities for new discoveries. Moreover, continuous feedback from integrated diagnostics supports education. Access to data from integrated diagnostics can facilitate shared decision making among patients and their clinicians and promote health equity by enabling personalized precision cancer care, she said.
For personalized treatment, Elenitoba-Johnson said that no single entity or clinical domain has all the tools or expertise necessary to measure, abstract, and interpret all the necessary data. To address this gap, integrated data science is “leveraging platforms that horizontally integrate information from disparate sources in a standardized fashion,” he noted (see Figure 1).
As an example, Elenitoba-Johnson described the interconnected Honest Broker for BioInformatics Technology platform to integrate digital pathology data from multiple sources and support its use for clinical, research, and educational purposes at MSK. He noted that MSK has digitally archived more than 5 million pathology slides to enable reliable retrieval of imaging data and the development of enhanced reporting through detailed digital annotation (Roth et al., 2021).
Another example of an integrated information system he described is Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT®),4
___________________
3 A tumor board is “a treatment planning process in which a group of cancer doctors and other health care specialists meet regularly to review and discuss new and complex cancer cases. The goal of a tumor board review is to decide as a group on the best treatment plan for a patient. These meetings can involve specialists from many areas of health care, including medical oncologists, radiation oncologists, surgeons, pathologists, radiologists, genetics experts, nurses, physical therapists, and social workers.” See https://www.cancer.gov/publications/dictionaries/cancer-terms/def/tumor-board-review (accessed December 27, 2023).
4 See https://www.mskcc.org/msk-impact (accessed August 30, 2023).
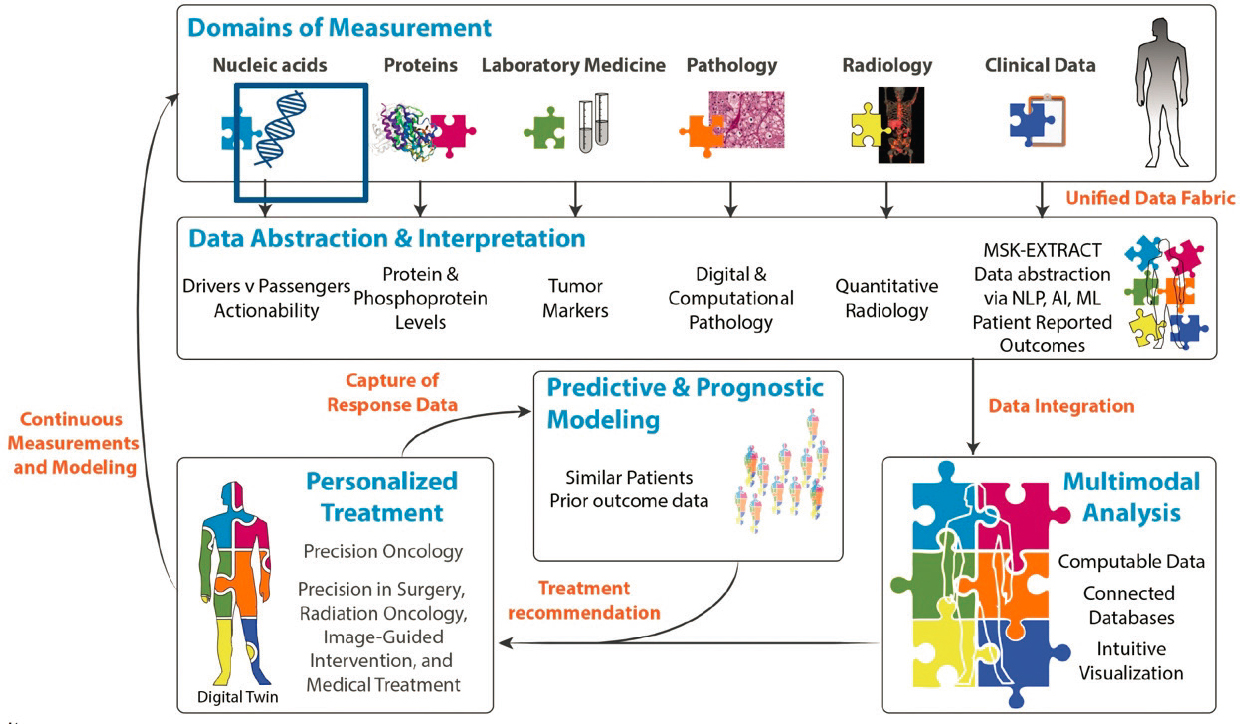
SOURCES: Kojo Elenitoba-Johnson and Hedvig Hricak presentations, March 6, 2023, and N. Schulz, L. Mellinghoff, and H. Hricak. 2017. Physical Sciences into Medicine Memorial Sloan Kettering Strategic Workshop.
a tumor gene sequencing system targeting 505 genes to create a molecular tumor tissue profile (Zehir et al., 2017). To support the standardization, interpretation, and dissemination of the clinical and genomic tumor data generated,5 a precision oncology knowledge base called “OncoKB” was developed (Chakravarty et al., 2017). Elenitoba-Johnson explained that MSK provides every patient with a personalized, comprehensive diagnostic assessment based on their MSK-IMPACT tumor testing, which includes “information about actionable mutations that are prognostically relevant.”
An Expanding Role for AI
Elenitoba-Johnson shared several examples of how AI is being leveraged to enhance the capabilities of integrated diagnostics in precision oncology care. As noted, MSK has digitized more than 5 million pathology slides, which can be used to train machine learning (ML) algorithms to identify cancer in tissue samples, often with improved accuracy compared to human review. He shared other examples of how AI is being used to improve data integration, such as studies using ML to improve risk stratification of patients with high-grade serous ovarian cancer (Boehm et al., 2022a) and prediction of response to immunotherapy in patients with non-small-cell lung cancer (Vanguri et al., 2022). He noted that digital pathology platforms also enable remote diagnostics, which can increase capacity, promote health equity, and, when implemented at scale, could potentially be used to improve cancer care and outcomes at a population level.
But Elenitoba-Johnson also cautioned that “AI is not infallible intelligence.” He explained that AI’s potential is affected by the quality of the datasets used to train the algorithms, automated discrimination associated with training datasets, human error, and the inability of algorithms to understand the context of the data (Chakravorti, 2022). He quoted four lessons learned from an analysis of the use of AI in the response to the COVID-19 pandemic (Chakravorti, 2022):
- Better ways to assemble comprehensive datasets and merge data from multiple sources are needed.
- A diversity of data sources are necessary.
- Aligned incentives can help ensure greater cooperation across teams and systems.
- International rules for data sharing are lacking.
___________________
5 See https://www.oncokb.org/ (accessed August 30, 2023).
To address these issues and advance the use of AI in biomedical research, the National Institutes of Health (NIH) Common Fund has launched the Bridge to Artificial Intelligence (Bridge2AI) program.6 Areas of focus include creating “flagship datasets” that are ethically sourced and adhere to the FAIR (findable, accessible, interoperable, and reusable) principles.7
Implementation Challenges
Hricak mentioned the exponential growth in diagnostic testing, but noted that testing tends to be siloed, which presents significant challenges for oncology clinicians in assimilating and interpreting the disparate diagnostic data for patient care. She underscored that the overall goal of an integrated data science approach to personalized treatment is to integrate all domains of measurement. However, challenges to achieving this goal range from data extraction and aggregation to integration and analysis.
Hricak said that data extraction challenges are substantial for pathology, radiology, and clinical data domains. Taking radiology data as an example, she noted that results reporting has remained essentially unchanged for well over a century and is still largely free text. Hricak explained that the lack of uniform, structured, and synoptic reporting8 leads to diagnostic uncertainty. Despite great interest in AI for tumor segmentation, feature extraction, and classification, this process remains largely manual. Manual segmentation, which identifies the spatial location of a tumor, is “the greatest roadblock to integrated diagnostics in both clinical and research workflows,” according to Hricak. She emphasized the need for “automated, algorithmic, validated tumor segmentation for every [body] site and every [imaging] modality.”
Hricak pointed to data governance and the culture around data sharing as additional challenges affecting the fields of radiology and pathology. Protections are needed to “prevent misuse and misinterpretation of data; protect interests of patients, faculty, multidisciplinary tumor boards, departments, and institutions; and ensure proper recognition of contributions,” noted Hricak, adding that it is also important to “be mindful of both scientific and business interests for all parties.” Hricak said that challenges for data integration and multimodal analysis include the need to develop new algorithms and identify new biomarkers for predictive and prognostic modeling.
___________________
6 See https://commonfund.nih.gov/bridge2ai (accessed May 26, 2023).
7 See https://www.go-fair.org/fair-principles/ (accessed December 27, 2023).
8 Synoptic reporting “is a method of clinical documentation that captures and displays specific data elements in a specific format.” See https://radiopaedia.org/articles/structured-reporting (accessed December 27, 2023).
Hricak stressed that the concept of integrated diagnostics is much more than a data aggregation dashboard; integrated diagnostics are an essential element of clinical decision support (NAM, 2017). Although both integrated diagnostics and clinical decision support tools are used with the goal of reducing medical errors, integrated diagnostics specifically focus on diagnostic errors (NASEM, 2015).
Until the goal of integrating all domains of measurement can be achieved, “every integration helps,” Hricak said. For example, when ruling out bone metastasis in patients with multiple myeloma, 18F-Fluorodeoxyglucose (FDG)–positron emission tomography (PET) results were negative in 11 percent of patients who had magnetic resonance imaging (MRI) findings of bony lesions in the spine (Rasche et al., 2017). Thus, integrating MRI and FDG-PET results can help ensure that patients who have metastatic tumors are not missed. Referencing another study that examined the use of integrated multiplatform omics tests and imaging to identify potential mechanisms of therapeutic response and resistance in metastatic cancers (Johnson et al., 2022), Hricak pointed out that these data were manually curated by 20 people over nearly a year. The study authors noted that further development of methodology for integrative analysis would support broad implementation in both research and clinical practice. She concluded that making integrated diagnostics a reality requires vision, courage, agility, collaboration, and perseverance.
Efforts to Improve Interoperability
Travis Osterman, director of cancer clinical informatics at Vanderbilt-Ingram Cancer Center, likened integrated diagnostics to a pipeline being filled with information from different clinical disciplines. Interoperability, which connects all of these multidisciplinary segments of pipe together, requires not only establishing data standards but adopting and integrating them into clinical workflows, he said. Integrated diagnostics falter when gaps in interoperability result in suboptimal data handoffs. Osterman likened this to the pipeline ending and pouring data into a bucket that must then be hand carried to another pipeline. Text-based clinical notes, explained Osterman, are one example of data that end up in a “bucket” for manual transport.
Examples of Progress Toward Interoperability
Development of data standards
Osterman explained that some data in operative reports are entered as unstructured notes, which creates a gap in the integrated diagnostics pipeline. To address this, the American College of Surgeons Commission on Cancer requires that operative reports for select
oncologic surgeries meet technical and synoptic formatting standards.9 The incorporation of standardized data elements in structured fields will better enable the collection, retrieval, and sharing of surgical information, which Osterman called “a breakthrough for the field.” He added that standards for synoptic operative reports for additional oncologic surgeries are expected to be forthcoming. He explained that academic cancer centers accredited by the Commission on Cancer are now in the process of implementing the formatting standards, and such requirements by accreditors are one approach to facilitate adoption. Osterman pointed out that it remains to be determined how well these standards can be embedded into clinical workflows where surgeons often type or dictate their notes.
Alignment with clinical workflow
Osterman pointed out that standards for capturing cancer staging as structured data exist, although implementation has been generally poor. One study found that before launching an implementation initiative, only 20 percent of patient records contained such information (Emamekhoo et al., 2022). Osterman suggested that rates of structured staging at many cancer centers are even lower. Clinical staging is often recorded as text in the clinical note or assessment plan, even though most electronic health records (EHRs) have structured fields for it. The problem is that making the structured entry requires the oncologist to take extra time to launch a separate form, and Osterman said, “many don’t see the direct benefit to either them or their patients.” Thus, the standards exist, but the process to enter structured information does not fit into the workflow. He pointed out that the lack of such structured data negatively affects research, as it can be extremely challenging to identify patients with a particular stage of cancer for clinical studies. In the study cited, the solution deployed was to “force” oncologists to complete the structured staging fields before they can close out the clinical encounter, Osterman said. Although this approach increased entry of structured staging information to 90 percent, he called for better solutions that align with clinical workflows so clinicians can spend their time on patient care.
Adoption of standards
Recalling the early days of genomic testing, Osterman explained that results from outside laboratories were provided by fax and not only unstructured but often difficult to interpret. Reports were later provided in PDF, and many oncologists now access testing results from an outside vendor via an online portal. The challenge, he said, is that the results often stay in that portal and are not uploaded to the patient’s EHR. Using the pipeline imagery, Osterman pointed out that in these situations, the integrated diagnostics pipe-
___________________
9 See https://www.facs.org/media/avukq4nc/coc_standards_5_3_5_6_synoptic_operative_report_requirements.pdf (accessed May 26, 2023).
line ends, and it is not clear where to even find the bucket of data to carry to the next section of pipe.
To address this gap, the health level 7 (HL7) standards for reporting structured clinical genomics data (including cancer-based testing, as well as somatic, germline, and pharmacogenomic testing) were introduced in 2019.10 These standards were designed to align with the oncology workflow by making it easy to order genomic testing and receive the results. The HL7 reporting standards have been widely adopted by genomic testing laboratories, EHR vendors, and health care systems, Osterman said, and the number of institutions receiving genomic data from vendors in structured format continues to increase. Achieving interoperability means these data also are available for other diagnostic uses (e.g., clinical decision support, recommending clinical trials, population health).
Gaps and Opportunities for Improvement
Despite progress toward interoperability, Osterman said that ongoing work is needed to identify and fill gaps in data standards. One area for attention is disease status. Osterman pointed to the “minimal Common Oncology Data Elements” (mCODE) interoperable data standard. Built on Fast Healthcare Interoperability Resources (FHIR) standards for exchanging health care data, mCODE leverages existing data standards.11 Osterman said that it enables clinicians and researchers to understand and describe a patient’s trajectory across the cancer care continuum. He noted that the concept of disease status in mCODE is defined as a clinician’s qualitative judgment on the current trend of a patient’s cancer—whether it is stable, worsening (progressing), or improving (responding), based on one or more sources of clinical evidence. While criteria exist for assessing disease status, such as the Response Evaluation Criteria in Solid Tumors (RECIST),12 it was decided during the development of mCODE that no existing data standard for disease status fit all cases, and this definition was proposed. Osterman explained that adoption of data standards has been a persistent challenge, and it remains difficult to extract data on disease status from the EHR. He added that this is partly a workflow issue, and mCODE is working with EHR vendors to improve the structured capture of disease status information.
Osterman said that clinical trials are another area that requires attention. He identified a need to structure the inclusion and exclusion criteria in ClinicalTrials.gov such that they are interoperable with the patient data in the
___________________
10 See https://build.fhir.org/ig/HL7/genomics-reporting/ (accessed May 26, 2023).
11 See http://hl7.org/fhir/us/mcode/ (accessed May 26, 2023).
12 See https://recist.eortc.org/ (accessed August 31, 2023).
integrated diagnostics pipeline for clinical trial matching.13 Osterman also noted the opportunity to improve the interoperability of clinical protocols so they can readily be launched across multiple institutions, and the Clinical Trials Rapid Activation Consortium is working to make this a reality (Osterman et al., 2023).
Lawrence Shulman, professor of medicine and director of the Center for Global Cancer Medicine at the University of Pennsylvania Abramson Cancer Center, emphasized the importance of structuring patient-reported outcomes and the clinician’s overall assessment of a patient for inclusion in integrated diagnostics. Mia Levy, chief medical officer of Foundation Medicine, pointed out that it can take many years for consensus standards to be developed and then implemented by vendors, and standards-setting bodies are generally volunteer organizations. She inquired how the development of standards might be accelerated. Osterman highlighted the differences between the HL7’s approach in proposing standards for structured clinical genomics data and the approach taken by mCODE. The HL7 process was much slower because it was intended to be full scope at implementation and therefore necessarily considered all use cases during development, Osterman explained, whereas mCODE was developed based on two use cases, and it was understood from the start that it was a minimum set of common data elements that would not fully cover all use cases. He pointed out that the mCODE governance structure allows for rapid feedback from users and rapid addition of new data elements. Osterman added that mCODE follows the FHIR maturity model,14 and a committee of mCODE implementers works to ensure that new additions are compatible with current installations.
Several speakers discussed the transition from manual to automated annotation and segmentation of image-based diagnostics. Hricak described the extensive and time-consuming manual work needed to annotate images and the need to assemble very large datasets of annotated images for training AI algorithms. She emphasized the importance of collaboration among radiology, pathology, and AI experts in developing and validating these algorithms. Hricak also called for a standard lexicon to convey the degree of diagnostic certainty. Elenitoba-Johnson highlighted infrastructure challenges to achieving automated annotation, such as the U.S. health care system’s lack of any standardized mechanism to pay for the expertise and effort spent on manually annotating images at the scale required to train AI algorithms. “Somewhere in our health care delivery system, we have to account for the individuals who
___________________
13 See https://clinicaltrials.gov/about-site/about-ctg (accessed October 24, 2023).
14 See https://confluence.hl7.org/display/FHIR/FHIR+Maturity+Model (accessed January 25, 2024).
are going to do that work,” emphasized Elenitoba-Johnson, rather than relying on the current patchwork funding.
Levy agreed that lack of annotated images is a workflow problem, with no incentive for annotation to be completed. However, a drawback to incentivizing time spent for annotation could be that clinicians and health systems may be reluctant to lose reimbursement in favor of automated annotation. Sohrab Shah, chief of computational oncology and Nicholls-Biondi Chair of the department of epidemiology and biostatistics at MSK, noted that “manual, precise … and thorough segmentation is completely unscalable to the volume that is needed to actually train the models.” He raised the possibility that semiautomated, large-volume datasets, although potentially less accurate, could be an option for training algorithms. He wondered whether the decisions of radiologists and pathologists could be captured as they are reviewing images in their routine workflow and whether that information could be used for training. Hricak pointed to self-supervised algorithm training (discussed further below).
EFFORTS TO DEVELOP, IMPLEMENT, AND USE INTEGRATED DIAGNOSTICS
Representatives from academic medical centers, industry, and large health care organizations shared their perspectives and lessons learned from efforts to develop, implement, and use integrated diagnostics in clinical practice.
Perspectives from Academic Medical Centers
Overcoming Cultural Barriers to Change
Gabriel Krestin, emeritus professor of radiology at Erasmus Medical Center, University Medical Center (MC), Rotterdam, shared lessons learned from implementing integrated diagnostics at Erasmus MC. To provide context, Krestin explained that the Netherlands has a highly regulated national health care system. Erasmus MC is the largest academic medical center in the Netherlands, with patient care organized into seven divisions, one of which is Diagnostics and Advice, incorporating the departments of pathology, laboratory medicine, medical imaging, and pharmacy.
Krestin said that the vision for Diagnostics and Advice is to implement an integrated approach that encompassed three key steps: requests for diagnostic testing are sent to a front office; tests are performed in the appropriate department; and a comprehensive, integrated, final report is provided to the multidisciplinary care team. He said a survey found that hospital leadership and information technology (IT) vendors supported this approach, but there
was unexpected resistance from physicians. Krestin explained that pathologists expressed concerns about digitization turning pathology into a commodity and radiologists expressed concerns about additional workload and responsibilities. Referring clinicians expressed concerns about ceding control of diagnostic decision making for their patients.
Krestin explained that Erasmus MC began by implementing integrated diagnostics for several complex diseases for which the institution expected particular benefit—for patients and the health care system. However, referring physicians responded that integrated diagnostics were unnecessary, because they could consult clinical practice guidelines for complex diseases. Erasmus sought to understand how well guidelines were being followed for these types of diseases. Krestin cited a retrospective review of 604 cases in which patients had incidental adrenal findings; of these, less than 15 percent followed Erasmus guidelines that called for both imaging and biochemical workups (de Haan et al., 2019). Retrospective reviews of guideline adherence for other scenarios yielded similar results. Even with this evidence of limited guideline adherence, “we couldn’t convince the clinicians that integrated diagnostics would be the way to go forward,” said Krestin.
A second attempt at implementing integrated diagnostics was to launch a pilot study to demonstrate how integrated diagnostics could improve workflow by addressing discrepancies between radiology and pathology findings before presenting a patient’s case to the multidisciplinary tumor board. Krestin provided an example showing agreement in diagnosis between radiology and pathology in approximately 75 percent of 89 patients with suspected lung cancer. An integrated approach led to reconciliation of an additional 20 percent of patients (unpublished data). Krestin pointed out that despite these results, hesitancy about integrated diagnostics among referring physicians persisted.
Krestin underscored that the main barrier to implementing integrated diagnostics was cultural (i.e., resistance from physicians). He said the necessary elements for successful implementation include transitioning from a process of multiple sequential orders to order sets (combinations of tests that support the determination of a comprehensive final diagnosis); development of an order management system that generates the order sets and defines the materials needed for testing; and structured, integrated reports delivering the combined results. Krestin added that Erasmus MC is now looking to integrate clinical and omics data for use in predictive analytics.
Patient-Centric, Functional Alignment of Diagnostic and Administrative Components
“Integrated diagnostics is a functional alignment of the meaningful diagnostic and relevant administrative components for a specific patient,” said
Jochen Lennerz, medical director of the Center for Integrated Diagnostics (CID) and associate chief of pathology at Massachusetts General Hospital. “It is not just the integration of multiple modalities.”
Lennerz described the work of CID, which aims to bridge the gap between clinical research and clinical practice. CID integrates a research laboratory, a clinical laboratory certified under the Clinical Laboratory Improvement Amendments15 and approved by New York State, faculty in pathology, and faculty in pathology informatics. CID runs approximately 14,000 clinical tests per year, with a baseline battery of tests and an agile infrastructure that facilitates both bringing new tests on board (including adopting payer policies) and removing tests.
Achieving diagnostic quality requires aligning the diagnostic components within the health care ecosystem, Lennerz explained. He described diagnostic quality as the sum of the quality of the diagnostic test itself, plus the quality of the diagnostic procedure (i.e., the execution of the test), and the quality of the diagnostic service (Lennerz et al., 2023). The integration process for a quality diagnostic test involves assessing the technology, determining financial sustainability (i.e., reimbursement by payers), and integrating it at the relevant laboratory.
As an example of implementing integrated diagnostics in practice, Lennerz described the testing approach to determine eligibility for targeted therapy among patients with lung cancer. The process involves collection of a biopsy sample, diagnosis by frozen section and fine needle aspiration, next-generation sequencing of extracted DNA, reporting of the genotyping results to the specialty pharmacy for dispensing of medication, and initiation of targeted therapy, which he said would ideally occur within 48 hours. In the example of initiating osimertinib16 therapy for patients who have lung cancer with EGRF mutations,17 the median time from the biopsy order to initiating therapy in a rapid specialty pharmacy cohort was 5 days compared to around 40 days in a nonrapid cohort (Dagogo-Jack et al., 2023). The potential of integrated diagnostics, he said, is “to work as seamlessly as possible to achieve your intended outcome.”
___________________
15 See https://www.cms.gov/medicare/quality/clinical-laboratory-improvement-amendments (accessed January 30, 2024).
16 Osimertinib is a kinase inhibitor used to treat or inhibit the formation of non-small-cell lung cancer. See https://medlineplus.gov/druginfo/meds/a616005.html (accessed September 1, 2023).
17 EGRF stands for epidermal grown factor receptor, and a mutation in this protein can cause uncontrolled growth, leading to cancer. See https://www.lung.org/lung-health-diseases/lung-disease-lookup/lung-cancer/symptoms-diagnosis/biomarker-testing/egfr (accessed September 1, 2023).
Despite frequent discussion about eliminating silos within the data infrastructure, Lennerz said “in our experience, integrated diagnostics works best if you have the siloed infrastructure but align all the components, including the administrative components, in a way that [is] harmonized for a specific workflow.”
Lennerz highlighted a variety of integrated diagnostics initiatives at Massachusetts General Hospital, including a cross-discipline and multimodality AI initiative in which diagnostic computed tomography (CT) scans are used to predict the molecular findings after biopsy and then, in a continuous feedback loop, those findings inform a continuously learning radiology model (Lennerz et al., 2023). He noted that a primary aim is to expedite authorization procedures.
Lennerz summarized that integrated diagnostics at CID value “individuals and interactions over processes and tools, sustainability over quick wins, specific journeys rather than general application, payer operations in addition to innovation-driven funding streams, [and] patient centricity rather than solely a scholarly exercise.”
Integrated Diagnostics as the Fourth Revolution in Pathology
Manuel Salto-Tellez, professor of integrative pathology at the Institute for Cancer Research, London (ICR) and director of the Integrated Pathology Unit at ICR and the Royal Marsden Hospital, posited that integrated diagnostics are the “fourth revolution in pathology,” following the introduction of immunohistochemistry in the 1980s, molecular diagnostics in the 2000s, and AI pathology solutions in the late 2010s (Salto-Tellez et al., 2019). He observed that, even with ideal application of genomic analysis, the number of patients with cancer who benefit from genome-targeted therapy remains low (Marquart et al., 2018), and although the performance of AI for digital pathology has been shown to improve with algorithm training, it appears to have plateaued (Echle et al., 2020). In his view, the next leap in pathology will likely not be another disruptive technology but an approach to integrating information for diagnostics and discovery of complex biomarkers.
Salto-Tellez defined integrated diagnostics as an “amalgamation of multiple analytical modalities, with evolved information technology, applied to a defined patient cohort, and resulting in a synergistic effect in the clinical value of the diagnostic tools” (Messiou et al., 2023). Current models of multimodal integration incorporate three components aligned with this definition: data modalities (the “what”), ML and integration analysis (the “how”), and opportunities for precision health (the “why”), and he cited five examples (Acosta et al., 2022; Cui et al., 2023; He et al., 2023; Lipkova et al., 2022; Lippi and Plebani, 2020). He suggested that a hierarchy for integration exists, considering the relative importance and timing of the information. In addi-
tion to providing opportunities for precision health, these models show that multimodal data integration can also support research and development, citing two examples of studies of integrated diagnostic approaches to predicting response to therapy (Sammut et al., 2022; Vanguri et al., 2022). Salto-Tellez shared that the Royal Marsden Hospital and ICR have jointly launched the Integrated Discovery and Diagnostics initiative, which merges genomic, histologic, radiologic, and clinical data, health care economics data, and research and development data from clinical trials.
One key challenge for multimodal integration of data across clinical silos is the extent to which data need to be curated versus obtained in an automated fashion directly from the source, said Salto-Tellez. He noted that Royal Marsden Hospital and ICR approached 20 vendors to help address this challenge. Of the nine that responded, six said they could offer a holistic approach; they had higher technical capability and cost. Three vendors said they could not integrate everything but could offer a piecemeal process that could help (they had lower technical capability and cost).
Salto-Tellez also described work by a consortium of the Precision Medicine Centre of Excellence at Queen’s University Belfast, Roche, and Sonrai to improve early detection of colorectal cancer. The consortium has developed an integrated diagnostics workflow connecting the pathology and genomics workflows to support the research and development process. The informatics partner, Sonrai, has developed the data analytics algorithms that link the two workflows together at multiple junctures, such as one designed to optimize the selection of tumor material for molecular testing based on histologic staining and digital pathology and to automatically macrodissect the tumor material. Salto-Tellez noted that validation studies of the integrated algorithms are underway.
Improving Diagnostics Through Close Partnership of Radiology and Pathology
Mitchell Schnall, Eugene P. Pendergrass professor of radiology and chair of the department of radiology at the University of Pennsylvania Perelman School of Medicine, observed that diagnostics has long been part of the art of medicine, often requiring the clinician to gather information from multiple sources to reach a diagnosis. However, the exponentially expanding volume of diagnostic data makes it impractical for one individual to integrate all of these inputs. Schnall said that data science can be leveraged to integrate a patient’s diagnostic information and present it to the clinician to review and act on, similar to how the cockpit of a plane presents all the necessary information to a pilot (Schnall et al., 2023).
Schnall discussed using partnerships to address silos, emphasizing that a closer partnership is needed between the two largest diagnostic disciplines—
radiology and pathology. He described this as a “natural partnership” because they have complementary expertise and approaches and face similar clinical and operational challenges. For example, pathology brings expertise in biology, informatics, and quality systems to the partnership, Schnall explained, and radiology brings expertise in anatomy and physiology, data science and IT, and workflow.
Schnall outlined the University of Pennsylvania’s approach to establishing a closer partnership between radiology and pathology, highlighting strategies in five tactical areas:
- Joint research includes building a joint data science program focusing on image analytics and overall diagnostic analytics to improve efficiency, defining the value of diagnostic outcomes, and developing novel combined modality diagnostics.
- Harmonization of systems includes using a single medical record system, the same picture archiving and communication system, and developing diagnostic decision support.
- Education includes cross-department house staff rotations, combined modality fellowship training, and a joint informatics/innovation fellowship.
- Joint clinical activities include codevelopment of diagnostic pathways, joint diagnostic consultation services, and promoting interaction through colocation of select pathology and radiology services.
- Integrated activities include the management and communication of results; decision support; and IT, billing, and marketing activities.
Schnall also said that the University of Pennsylvania recently launched the Center for AI and Data Science for Integrated Diagnostics (AI2D). Codirected by faculty in radiology and pathology, AI2D focuses on addressing the challenges of integrated diagnostics. “Diagnostics is critical to precision medicine,” Schnall concluded, and “closer integration of radiology and pathology will improve the diagnostic process.”
Integrated Diagnostics for Earlier Detection of Cancer
Garry Gold, Stanford Medicine Professor of Radiology and Biomedical Imaging and chair of the Department of Radiology at Stanford University, discussed the wide-ranging applications of integrated diagnostic approaches to early detection of cancer, when treatment is more likely to be successful.
Gold first described several studies by the late Sam Gambhir, a founder of the Canary Center at Stanford for Cancer Early Detection and pioneer in the integration of radiation and pathology. In one example, Gambhir assessed
whether integrating FDG-PET imaging with assessment of circulating tumor cells (Nair et al., 2013) or circulating tumor microemboli (Carlsson et al., 2014)18 in patients with non-small-cell lung cancer could improve diagnostic accuracy over using radiology or pathology markers alone. Ghambir also initiated the Baseline Study,19 a longitudinal study collecting baseline clinical, imaging, molecular, and other data from 10,000 participants to characterize what “normal” values are and changes that occur in association with disease.
Gold highlighted a novel approach known as “theragnostics,” which combines diagnostic testing with therapy, such as the integration of diagnostic radiology and targeted molecular radiotherapy. The Canary Center is also developing new in vitro diagnostic (IVD) tests for early cancer detection, including the Exosome Total Isolation Chip, which can detect extracellular vesicles in clinical fluid samples (Liu et al., 2017); a magnetic wire inserted into a vein to capture circulating tumor cells that have been immunolabeled with magnetic particles (Vermesh et al., 2018); and an approach that correlates detection of volatile organic compounds in breath with PET-CT (Vermesh et al., 2022).
Gold highlighted wearable and in-home monitoring technologies as another opportunity for integrated diagnostics. One example of a wearable technology is a microneedle patch on the arm that integrates sampling and molecular testing to continuously monitor for proteins in interstitial fluid. Gold also noted that integrated diagnostics are being developed in other fields, such as combining neuroimaging and histopathology data to assess traumatic brain injury.
Achieving the Vision of Integrated Diagnostics
Several panelists discussed potential actions that could help realize the vision of integrated diagnostics in precision cancer care. “It’s early days in integrated diagnostics,” Salto-Tellez said, with a need for “significant and specific investment in this area.” Lennerz encouraged an increased focus on regulatory science and actively engaging with regulators on issues related to practical implementation (e.g., the data and performance metrics needed for radiologists to fully integrate AI) (Lennerz et al., 2022). Schnall stressed that broad collaboration among academia, industry, regulators, and payers is necessary
___________________
18 Circulating tumor cells are cells that have separated from primary or metastatic tumors. Circulating tumor microemboli are multicellular aggregates that include circulating tumor cells, and both of these can seed metastatic cancer in disparate sites in the body (Tao et al., 2022).
19 See https://medicine.stanford.edu/annual-report-2018/the-project-baseline-study.html (accessed September 1, 2023).
to achieve effective integrated diagnostics. Gold said the next generation of scientists and clinicians need to be able to communicate across disciplines and implement an integrated approach to diagnostics. “We won’t be able to truly integrate until we have people who understand the entire landscape,” he said.
Several participants discussed the challenges of integrating the vast volumes of data generated by consumer wearables and in-home monitoring technologies. Shah raised the issue of access to these data by hospital systems or clinicians, noting that wearables are often commercially developed, and the developers often have their own interests in the data collected. Gold agreed and noted the regulatory, payer, and patient privacy concerns to be addressed.
Perspectives from Industry
Opportunities to Leverage AI in Diagnosis
Dorin Comaniciu, senior vice president of Artificial Intelligence and Digital Innovation at Siemens Healthineers, discussed several opportunities to leverage AI for integration and use of the massive volumes of health data generated by medical technologies and devices. He said that computational power and bandwidth for information exchange are increasing exponentially, including low-cost supercomputing power available at the point of care.
Comaniciu said that one approach to handling the ever-increasing volume of data is AI self-supervised learning.20 He cited a study of self-supervised learning using 100 million medical images, which found that it resulted in improved accuracy, more robust results, accelerated training, and improved scalability (Ghesu et al., 2022). Comaniciu noted that it would take 35 years for one person to look at each of the 100 million images for 10 seconds.
The results of self-supervised learning would need to feed into a harmonized user interface, which Comaniciu referred to as a “diagnostic cockpit,” with access to clinical information, the results of AI analyses, advanced data visualization, actionable reporting, and research and innovation. Comaniciu pointed out that structuring data has been the general approach to managing complex clinical data so it can be integrated. However, AI has the capability to shift the model from one in which the clinicians sort through structured data to one in which clinicians ask questions (comparable to Internet searching). He said large language models will support connectivity and access, provide interpretation support, and aid in synthesizing information. In addition,
___________________
20 Self-supervised learning is an ML technique that teaches a model to predict hidden parts of an input using other parts of the input that are visible to the model. It can be used to perform tasks, such as image comprehension and object detection (Rani et al., 2023).
this workspace could incorporate virtual assistants capable of responding to questions about the patient. To illustrate this, Comaniciu shared a video of a conversation with an AI radiologist assistant that responded to successive questions about the patient’s testing and findings.
Comaniciu presented several examples of AI-powered diagnostics for oncology, such as cancer risk prediction, brain tumor analysis and metastasis digitation and tracking, lung cancer screening via chest CT, assessment of pulmonary lesions via chest X-ray, breast cancer screening, prostate MRI, and analysis of cardiotoxicity. He also highlighted AI tumor fingerprinting, an approach under development to create a digital biopsy of molecular changes in tumors by integrating digital pathology, proteomic, metabolomics, and genomic profiling. Another fingerprinting approach is using an image-based deep neural network to optimize radiation therapy by stratifying patients as responders or nonresponders to guide individualized dosing (Lou et al., 2019; Randall et al., 2023).
Looking to the future, Comaniciu anticipated patients would have a “health digital twin,” which he characterized as a “lifelong, personalized, physiological model that is updated with each scan exam.” Simulations with a digital twin could inform treatment decisions, for example, or guide individualized preventive actions, he noted.
Building in Feedback Loops to Improve Quality of Care
Torbjörn Kronander, president and chief executive officer of Sectra AB, compared diagnostics to telecommunications in that both must detect signals in noise. Diagnostic testing detects signals of disease.
Kronander outlined some of the key features of IT systems for health environments. All relevant data should be available in the same place at the same time because clinicians do not have time to go back and forth among systems to find patient information, he said. Deep integration with AI is needed, and “AI should be applied both to radiology and pathology at the same time,” he said. The challenge, however, is that these data often reside in different systems. Uniform user interfaces can increase patient throughput, he noted, because moving to a user interface with a different format requires time to “reconfigure your brain” and adds “mouse miles” (computer mouse movements needed to engage with interfaces). Kronander added that unified tumor boards would help ensure that patient treatment is not delayed while discordant conclusions from pathologists and radiologists are resolved. Early identification and resolution of discordance among specialties can help the tumor board to work more efficiently, he noted.
Kronander explained that Sectra is working to integrate “radiology, pathology, and AI in one single IT system.” There is close integration with
information in the patient’s EHR, and Sectra is developing prototype structured reports that combine radiology, pathology, and concordance activities in one report sent to the EHR (he noted that the Sectra system currently only interfaces with the Epic EHR system). He added that Sectra is also collaborating with the University of Pennsylvania School of Medicine to develop a single user interface that incorporates pathology, radiology, and genomics.
Sectra’s approach to integrated diagnostics includes radiology, pathology, and omics data combined with current population health and probability data, Kronander said. He noted that probability data are often overlooked but are important for establishing a diagnosis. He explained that a key element of the Sectra integrated diagnostics model is feedback loops to support a learning system. Data from the structured diagnostic report in the EHR (where the final interpretation/outcome is entered) are also fed back to radiology, pathology, and omics functions and used to update population health and probability data.
The vision for integrated diagnostics, Kronander said, is “faster and better diagnosis, improved patient care, built in feedback loops [for] improving quality of health care, and at lower cost and less complexity.” He referred to Gambhir’s prediction that “[t]oday’s radiologists and pathologists will be replaced by diagnosticians plus AI” (Gambhir, 2018). Achieving this vision, Kronander said, will require “common basic training in decision theory for radiologists and pathologists.”
Kronander said that changes in reimbursement are also needed to achieve the vision of integrated diagnostics. He suggested that payers should reimburse clinicians for making the diagnosis rather than conducting specific diagnostic procedures. He acknowledged that such changes take time and observed that the U.S. system of reimbursement can be a barrier to the timely advancement of health care practice. Elenitoba-Johnson added that one challenge is coming to an agreement regarding “who is going to do the work and who is going to pay for it.” He emphasized that incentives need to be aligned to promote the adoption of integrated diagnostics into clinical practice.
Harnessing the Power of Data for Diagnosticians, Clinicians, and Patients
Nick Trentadue, director of laboratory and diagnostics informatics at Epic, identified some settings in which patient health care data are generated (see Figure 2). Traditionally, test results would be reported to the patient’s clinician, who would share them with the patient and discuss treatment options. Today, the results are also provided immediately to patients. Trentadue shared a screenshot of an actual report received by a patient awaiting cancer screening results. These reports, written by and for medical professionals, can be confusing and frightening to patients, he cautioned. Trentadue highlighted the need for tools that can better convey test results to patients.
SOURCE: Nick Trentadue presentation, March 6, 2023.
Trentadue said integrated diagnostics need to be considered from three primary perspectives: diagnosticians, treating clinicians, and patients. For diagnosticians, a key element supporting the diagnostic process is the bidirectional exchange of interoperable data across systems. Interoperability requires that health data vendors adhere to industry standards for exchanging health information across systems (e.g., the HL7 standards for structured clinical genomics data discussed by Osterman). For treating clinicians, combining the results from the diagnosticians with AI and ML creates models that can support clinical decision making and the development of care plans that meet individual patient needs. For patients, Trentadue said that timely updates via the patient portal should leverage integrated data to provide results (e.g., laboratory, pathology, radiology), easily understood interpretations, and next steps for patient care.
Trentadue emphasized that for integrated diagnostics to optimize the power of data, the information should be discrete and actionable and follow standards that enable interoperability and support bidirectional flow among the care team, regardless of data system: this ensures that “the right person, at the right time, has all the appropriate information for diagnosis as well as for treatment.” Trentadue also emphasized the importance of ontology21 when developing ML models, because a lack of clear ontology could lead to unexpected outcomes from the models, and actionable decision support, in which diagnosticians, treating clinicians, and patients have the information they need, readily available, to support decision making.
Integrating Patient Goals and Treatment Outcomes Data
Aanand Naik, professor and the Nancy & Vincent Guinee Chair of Geriatrics at the University of Texas Health Science Center, Houston, School of Public Health and Consortium on Aging, highlighted the need to incorporate patient goals and health outcomes (including the potential for adverse effects) into integrated diagnostics. Kronander added that continuous, intensive monitoring of treatment outcomes is needed because of the increasing number of treatment options. If one therapy is not achieving an expected outcome, then a change may be necessary. This type of monitoring will also drive the use of diagnostics, he said. Comaniciu noted that it is often not possible to determine the best treatment approach for a given patient based on available guidelines. He anticipated that computer simulations could eventually be able to rank the top treatment choices for a patient to consider.
___________________
21 An ontology, in the context of computer science, describes the relevant concepts, relationships, and specifications that are important for modeling a particular domain (Gruber, 2016).
Transparency of AI Models
Osterman said clinicians are reluctant to base their decision making on recommendations from closed-box AI models. Trentadue responded that transparency is essential when presenting information gleaned from AI, ML, or algorithms. Kronander noted the ongoing research on explainable AI,22 emphasizing that AI should not be fully independent; humans need to be in the loop to identify instances when AI does not work.
Perspectives from Large Health Care Organizations
Precision Oncology in VA Medical Centers
Gil Alterovitz, director of the National Artificial Intelligence Institute (NAII) of the U.S. Department of Veterans Affairs (VA), said the vision of NAII is “to lead the way in trustworthy AI” with the goal of “ensuring the health and well-being of our veterans.” NAII is building an organization focusing on AI research, development, and training through a network of AI sites to “pilot, iterate, and scale” AI initiatives within the VA, Alterovitz said; it has four sites and several pilot programs.
The VA is well situated to take the lead in advancing AI in health care, Alterovitz said. The VA is the largest U.S. integrated health care system, serving more than 9 million patients. The VA has a repository of more than 10 billion medical images and a database of more than 800,000 genomes linked to medical records. In addition, the VA has a broad reach, with more than 1,200 facilities across all U.S. states and territories. He added that most clinicians in the United States have undertaken part of their training at a VA facility.
Alterovtiz said the “VA is the largest provider of oncology services in the country, making oncology care a priority.” He shared several examples of how the VA is bringing advances in technology and precision care to veterans. As part of the federal Cancer Moonshot initiative, the VA has established a public–private partnership with IBM to enable precision care for patients with stage 4 cancer who have exhausted available treatment options. Researchers use IBM Watson AI to analyze a patient’s tumor for mutations to identify potential options for targeted therapy. Alterovitz said that this approach has expanded patient access to precision oncology therapy; more than one-third of the tumor samples are from patients who live in rural areas, where access to such therapies may be limited.
___________________
22 See https://en.wikipedia.org/wiki/Explainable_artificial_intelligence (accessed January 25, 2024).
Another example is the VA’s Lung Precision Oncology Program,23 a network of 23 hub locations working to “increase access to screening and improve early detection” using data analytics to evaluate access and quality. Alterovitz noted that more than 5,000 veterans received molecular testing through this program as of May 2021.
For VA research programs, such as the Lung Precision Oncology Program, Alterovitz said participants undergo a consent process to share their data and a terms and conditions process by which veterans agree that their data can be used for operational purposes. Depending on who will be using the data and how, data use agreements and contracts may also ensure patient privacy and data security in accordance with patient preferences. He noted that it is better to consider potential data uses and secure patient consent at the beginning rather than having to reconsent patients later for additional uses.
Alterovitz also discussed the use of AI-assisted screening to detect precancerous polyps during routine colonoscopy. Alterovitz noted that “every 1 percent increase in the precancerous polyp detection rate reduces future risk of death by 5 percent.” Studies of this AI-assisted colorectal cancer screening tool have demonstrated a 14 percent increase in the rate of precancerous polyp detection.
Alterovitz noted that the VA has an app store with a range of apps to help veterans access their medical information, receive care (e.g., testing, medications), or find appropriate clinical trials based on information in their medical record.24 In closing, he said that the VA is looking for opportunities to leverage its data gathering and analysis capacity in collaborations that advance the development and use of AI in precision cancer care.
Precision Oncology at University of California Health
Atul Butte, the Priscilla Chan and Mark Zuckerberg Distinguished Professor and director of the Baker Computational Health Sciences Institute at the University of California, San Francisco, and chief data scientist for University of California Health (UCH), provided an overview of UCH. The University of California (UC) system has more than 227,000 employees and 280,000 students per year across its 10 campuses. UCH incorporates UC’s 6 medical schools and 14 other health professional schools (nursing, pharmacy, veterinary, dental, and public health) and includes five National Cancer Institute (NCI)-designated comprehensive cancer centers and five institutes funded by the NIH National Center for Advancing Translational Sciences (NCATS) Clinical and Translational Science Award program. UCH employs
___________________
23 See https://www.research.va.gov/programs/pop/lpop.cfm (accessed September 1, 2023).
24 See https://mobile.va.gov/appstore (accessed May 26, 2023).
approximately 5,000 faculty physicians and 12,000 nurses and trains half the medical students and residents in California. It receives $2 billion in funding from NIH and brings in more than $16.5 billion in clinical operating revenue.
Butte said one original goal was to transform UCH into a single accountable care organization, facilitated by UC’s approach to data warehousing. The UCH-wide central data warehouse facilitates broad access to deidentified, structured clinical data from across all six academic health centers. A warehouse is still maintained at each of the six academic medical centers, which facilitates access to “deidentified, limited, and identified structured and unstructured clinical data” and additional information (e.g., images, genomes, notes). The central database has longitudinal, structured data going back to 2012. This includes data from 437 million patient encounters, 1.17 billion procedures, 1.5 billion medication orders, 48 million device uses, and 5.2 billion laboratory tests and vital signs, from nearly 9.2 million people. Data are merged with California state data and the California death index, Butte noted.
More than 100,000 patients with cancer receive care at a UCH comprehensive cancer center each year, noted Butte. The same UC-wide data warehouse contains demographic information, including information on a patient’s cancer diagnosis, insurance coverage and social risk, and more than 32,000 cancer genomic reports. Butte said these data can be queried to address a wide range of questions, such as whether patients who have a particular type of cancer with a specific gene mutation are receiving appropriate and equitable cancer treatment.
Butte explained that the UCH academic medical centers use the Observational Medical Outcomes Partnership Common Data Model. He noted that a systemwide committee meets every 2–4 weeks to harmonize data elements. Certain analyses require curation of unstructured clinical notes and data in the warehouses. Not having thousands of curators available, Butte described how UCH is looking at using an AI chatbot to curate deidentified clinical notes. He described one situation in which ChatGPT25 was asked to summarize all cancer biomarkers in a complicated deidentified clinical note. The AI system was able to find and summarize data for all of the biomarkers, including one that the UCH oncology curators had missed.
UC researchers (including graduate students) who want access to UCH-wide data first write and optimize their queries in their campus system and then use a cloud-based tool to run the queries in the central UCH-wide warehouse. The goal is to facilitate “safe, respectful, regulated, responsible use of clinical data,” explained Butte. He said that data in the UCH-wide central data ware-
___________________
25 ChatGPT stands for Chat Generative Pre-trained Transformer and was created by OpenAI. It is a large language model–based chatbot. See https://openai.com/blog/chatgpt (accessed September 1, 2023).
house are deidentified and can therefore be shared in compliance with the Privacy Rule26 promulgated under the Health Insurance Portability and Accountability Act (HIPAA), and no institutional review board (IRB) approval is needed to conduct studies with these data. UCH has also determined that tumor mutation data are nonidentifiable and can be shared. He also noted that every patient is required to sign a terms and conditions of service document, which outlines how the university may, in compliance with HIPAA, use their data. A governance process is also in place to review external requests for the use of UCH data.
Training AI requires vast amounts of data, but competition among health systems is a barrier to data sharing across the country, Butte noted. While acknowledging the challenges, Butte contended that the goal is achievable, offering the NCATS/NIH National COVID Cohort Collaborative27 as an example. Butte described how NCATS successfully motivated more than 100 health institutions to contribute deidentified data for all of their patients with COVID-19 to a central, third-party repository to support research on the disease. Data are publicly accessible through dashboards. Butte emphasized that this collaborative has led to nearly 500 data projects and 53 publications thus far. Butte pointed out that NCI does not require similar data sharing among their funded comprehensive cancer centers, and he advocated for the creation of a common data warehouse that could enable the development of precision medicine tools for patients with cancer.
Butte said that interoperability of health data is achievable, and a business need might drive the implementation of data sharing, but a culture that supports data sharing is also needed. Converting unstructured to structured data remains a challenge, but there is a potential role for AI. Other challenges noted by Butte include ongoing resistance to cross-campus clinical trials despite incentives and central IRBs and a lack of precision cancer care options for patients.
IMPROVING EVIDENCE GENERATION FOR INTEGRATED DIAGNOSTICS
Many speakers discussed approaches to generating evidence to inform and support evaluating, implementing, and scaling integrated diagnostics.
___________________
26 The Privacy Rule, promulgated under the Health Insurance Portability and Accountability Act of 1996 (HIPAA), establishes national standards to protect individuals’ medical records and other individually identifiable health information (collectively defined as “protected health information”) and applies to health plans, health care clearinghouses, and those health care providers that conduct certain health care transactions electronically (see https://www.hhs.gov/hipaa/for-professionals/privacy/index.html).
27 The National COVID Cohort Collaborative is also known as N3C. See https://ncats.nih.gov/n3c (accessed September 1, 2023).
A Cancer Risk Management Learning System for Evidence Generation
Levy shared opportunities to leverage a learning health care system to evaluate integrated diagnostics. In such a system, she explained, local clinical guidance is developed based on national guidelines and scientific literature and implemented in the health care delivery system. Outcomes data generated as part of routine care are analyzed and used to inform updates to the local guidelines. Information also feeds back to research and publication in the scientific literature. This cycle results in iterative improvement to the care delivery system.
Levy described Rush University’s experience implementing a learning health care system for breast cancer risk management to address the challenges of evidence generation for breast cancer screening. Levy noted that the patient population eligible for breast cancer screening is large and diverse. While there are conflicting clinical practice guidelines for breast cancer screening, all guidelines recommend that screening begin with mammography. Mammography is highly regulated, and the use of structured mammography and other screening data documentation in the EHR has been increasing, Levy noted. There are also well-defined outcome measures for screening that can be extracted from the EHR. Although these could be used to evaluate new screening methods in prospective randomized trials, Levy pointed out the many challenges, from ensuring adequate patient accrual to the rapid evolution of screening technologies, and suggested a learning health care system could help generate evidence to evaluate integrated diagnostics.
In support of this model, Levy said the Rush system has structured data based on the Breast Imaging Reporting and Data System from more than 500,000 screening and diagnostic breast imaging studies dating from 2008. These data are from a diverse patient population (less than 50 percent White) and span a wide range of ages (from under 30 to more than 90, with most data from individuals aged 40–70). The amount and types of structured data available continue to increase, including breast density data (available since 2015) and cancer risk assessment information (from 2020 onward). “As our structured data has grown, so have our efforts to leverage this data to learn more and more from the experiences of every patient,” Levy said.
Levy outlined the elements of Rush’s breast cancer risk management learning health care system: implementing new clinical guidelines for risk assessment, risk-based supplemental screening, and risk management; implementing risk assessment tools, reporting, and patient navigation; and analyzing guideline uptake, distribution of risk, and automated calculation of cancer detection rates from EHR data (see Figure 3).
Levy noted that “breast screening is not a moment in time.” Integrated diagnostics need to account for the patient’s longitudinal screening experience (e.g., comparing new images to prior images). Furthermore, integrated
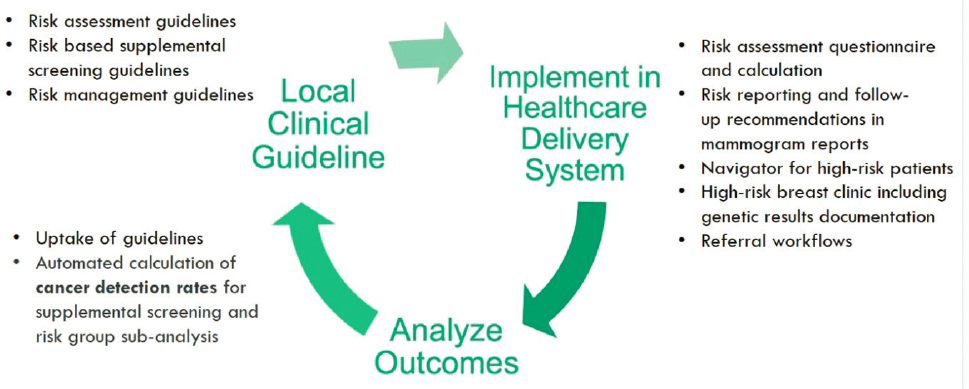
SOURCE: Mia Levy presentation, March 6, 2023.
diagnostics need to extend beyond test interpretation to include informing the next steps in patient management, she emphasized.
Levy explained that the risk management system is supported by clinical and diagnostic systems integrated into the EHR. She highlighted the use of screening test order sets that a clinician selects based on patient characteristics, including breast cancer risk factors, such as breast density and genetics. The next steps in the diagnostic pathway are driven by the breast imaging center, she said. Other integrated clinical systems include cancer staging documentation, high-risk clinic documentation, and risk assessment calculators. Diagnostics systems include documentation systems for radiology, structured genetics and pathology results, and patient outreach documentation and worklist management. Levy noted that most of the analytics needed for the learning health care system are not yet available from vendors and are being developed in house. These include analytics dashboards, quality measure analytics, and population analytics.
Over the first 3 years of the program, Levy said that about 7 percent of the total population had a personal history of breast cancer, and about two-thirds of those were eligible for supplemental breast MRI according to practice guidelines. They now are assessing what percentage of eligible patients underwent the supplemental breast MRI, she said.
Levy explained that most patients (84 percent) were eligible to undergo a cancer risk assessment. About 10 percent of those who did so were identified as high risk and therefore eligible for a supplemental MRI. The majority of patients were at average risk or declined the risk assessment. Those with dense breasts were eligible for supplemental automated breast ultrasound in addition to standard mammography screening. Levy said uptake was approximately 40 percent among patients for whom supplemental ultrasound was recommended, noting that this is less than ideal. She added that payer coverage of the supplemental testing significantly influences uptake.
Levy emphasized the need for training datasets to be representative of the population in which the technology is intended to be used. She said that the risk classification system was found to underestimate the risk of breast cancer among Black women (compared to White and Hispanic women) as a result of how the algorithm was initially trained. Levy also cautioned that an algorithm developed in one context can perform quite differently when applied in a different population, which underscores the importance of ensuring that training algorithms are drawn from diverse, representative datasets. Levy acknowledged the significant challenge of training AI algorithms to perform consistently when moving from local to regional or broader use, given the great population diversity across this country and the world.
In closing, Levy said, “Integrated diagnostics evaluation can be enabled by learning health care systems implemented in health care delivery systems,”
adding that the Rush experience demonstrated “the feasibility of this approach for evidence dissemination and evidence generation for cancer screening.”
Building the Evidence Generation Infrastructure to Scale Precision Diagnostics
One challenge of precision diagnostics is advancing a technology born of “great science” to a product that can actually be used at scale, said Bradford Hirsch, head of Product and Implementation at Verily/Alphabet. He described two examples of taking precision diagnostics to scale from outside the field of oncology.
The first example was real-time screening for diabetic retinopathy using a deep learning system (Ruamviboonsuk et al., 2022). To bring it to scale, Verily first had to build a camera that could be used by clinicians globally to reliably capture images of the retina. Hirsch said that the camera was built by a multidisciplinary team that understood the relevant regulatory and reimbursement pathways. The camera can autonomously identify the retina in approximately 98.5 percent of patients, capture an image, and transmit it to the cloud for AI image assessment.
In a second example, he said that Verily designed a “study watch” for the collection of digital biomarker data from patients with Parkinson’s disease (Burq et al., 2022). Hirsch explained that despite many consumer wearables already available, it is generally not possible to access primary data from other devices—sensors and algorithms are frequently changed, for example—or customize the user experience. The watch developed by Verily takes remote measurements of motor function of individuals with Parkinson’s by alerting the wearer to complete a timed motor task and report the outcome through a survey on the device.
Hirsch explained that after product approval, there is a “data void,” so phase 4 postmarketing studies may be needed to gather additional data. This data void presents a challenge for precision diagnostics developers looking to take products to scale in clinical care. Verily is working to develop an evidence generation infrastructure with better continuous data collection after approval, closing the data gap between data collected during clinical research and data collected as part of clinical care.
Hirsch explained that Verily has restructured, bringing its research and care teams together under the same leadership team. Merging product development and implementation datasets can facilitate the creation of “products that can actually be used in practice,” Hirsch said.
Hirsch outlined five key elements of Verily’s approach to improve evidence generation:
- Data-driven identification, recruitment, and enrollment of target populations in Verily research and care projects (e.g., using social media, information from payers).
- Informed participant consent and permission processes, which help to automate research activities such as recruitment or data aggregation. Hirsch explained that participants consent to specific elements of the project, for a specific time period, and also provide permission to be contacted again.
- Participant voice at the center. Participants are engaged “in a tactical, tangible way” through regular patient-reported outcome measures and other approaches to data collection, Hirsch said. Verily has a 75-person user experience team focused on motivating participants to take part in longitudinal data sharing.
- Augmentation with novel passive tools. Hirsch noted that replicating elements of surveys, such as the Eastern Cooperative Oncology Group or Karnofsky performance status scales,28 in consumer-facing technologies might be straightforward, but much effort is required for validation. For example, the watch’s virtual motor exam in the Parkinson’s disease study was compared to office-based motor assessments.
- Merging data generated with secondary datasets (e.g., clinical trials, EHRs, administrative claims data, or biology research) can enable “truly differentiated, actionable, regulatory-grade insights,” Hirsch said.
Hirsch stressed that the infrastructure for precision devices requires proactive monitoring of the real-world performance of AI-based tools that is intentional and considered in advance of implementation. Data collection can occur as part of routine clinical care, with the potential to aggregate data across devices and share data. Moreover, real-world performance data also enables continuous learning.
Opportunities to Leverage EHRs for Clinical Evidence Generation
Diagnostic testing affects all aspects of medical care, from screening and diagnosis to selection of treatment, follow-up, and surveillance, explained Neal Meropol, vice president of Research Oncology at Flatiron Health and chair of
___________________
28 The Eastern Cooperative Oncology Group performance status scale helps define a patient’s overall ability to function after treatment. The Karnofsky Performance Status scale seeks to determine a patient’s impairment after treatment. See https://ecog-acrin.org/resources/ecog-performance-status/ and http://www.npcrc.org/files/news/karnofsky_performance_scale.pdf (accessed September 1, 2023).
the NCI Director’s Clinical Trials and Translational Research Advisory Committee. In using diagnostics, clinicians need to understand test performance characteristics, including the test’s sensitivity, specificity, positive predictive value, and negative predictive value;29 the interpretability and actionability of the results; and the cost and convenience for the patient.
Meropol reviewed a few of the challenges to evidence generation for integrated diagnostics. Generally, the level of investment in evidence generation for diagnostics is lower than for drugs. In addition, technological advances are driving the development of numerous, increasingly complex diagnostic tools. As a result, clinicians are often faced with numerous testing options, many with inadequate clinical guidance, leaving them uncertain about what test to order or how to interpret the results.
He said that EHRs provide opportunities to gather evidence for integrated diagnostics, although they were not designed for research. EHRs are “a rich source of patient-level data” from multiple data sources, said Meropol. Much of the information in EHRs is digitized and available in real time. EHRs are embedded in the point-of-care workflow, which affords opportunities to provide clinical decision support in association with EHR data. Furthermore, data from EHRs tend to be more representative of the general population than data from clinical trials, which are from enrolled cohorts that trend toward “younger, healthier, Whiter, richer patients than the typical patient with cancer nationwide or worldwide,” he said. Meropol added that the U.S. Food and Drug Administration (FDA) has issued guidance addressing key considerations for using real-world data for clinical research and regulatory decision making (e.g., data quality, analytic approaches, fitness for purpose).30
EHRs can be a platform for integrating evidence both from within and outside the EHR system, observed Meropol. This includes diagnostic testing reports, pathology and radiology images, genomic data, patient-reported data and data from wearables, insurance claims data, information on social determinants of health, and mortality data. He explained that mortality data are often missing or inaccurate in EHRs, but it is now possible to link to population registries to provide more accurate mortality data.
Meropol said the key capabilities for the use of EHRs to generate evidence include a “privacy framework that respects patients”; linkages with outside data sources, which might require tokenization of data; curation of both structured
___________________
29 Sensitivity is the ability to identify a true positive, and positive predictive value is probability that a patient does have the disease if tested positive. Specificity is the ability to identify a true negative, and the negative predictive value is the probability that a patient does not have the disease if tested negative (Parikh et al., 2008).
30 See https://www.fda.gov/science-research/science-and-research-special-topics/real-world-evidence (accessed May 26, 2023).
and unstructured data; “careful application of ML and natural language processing to avoid misleading, inaccurate, or biased conclusions;” real-time data; and “context-specific characterization of data quality and fitness for purpose.”
Meropol emphasized the need for use cases and pointed out that not all data in the EHR are fit for purpose to address evidence gaps for integrated diagnostics, and information routinely collected in the course of clinical practice may be insufficient to address research questions. Passively collected data in EHRs may be characterized by missingness, confounding biases, selection biases, exposure variability, and variability in documented outcomes. One approach is to augment routinely-collected data with prespecified intentionally-collected data in the EHR to support prospective observational research studies (e.g., non-routine tests, testing at specific times or intervals, patient-reported outcomes).
Meropol described a case study in which Flatiron Health and collaborators used centralized, intentional EHR-based data collection and processing to support a prospective, observational study among patients with non-small-cell lung cancer.31 Circulating tumor DNA was obtained at prespecified time points to generate a dataset of integrated evidence that included genomic data, clinical data, and digital pathology, he said. One analysis, for example, compared the sensitivity of tissue-based genomic profiling to blood-based genomic profiling (Schwartzberg et al., 2022).
Meropol described the operational model implemented by Flatiron Health for gathering data from the EHR for prospective evidence generation. Data for research in the EHR may consist of routinely collected structured data (e.g., vital signs, laboratory tests, drugs), intentionally-collected structured data (not part of routine care but part of the routine workflow for research), and unstructured data (e.g., pathology reports, clinician notes). These data are transmitted from the EHR to a data warehouse through automated processes, with unstructured data first being processed by both humans and trained AI models. Data may then be linked to external sources of data, and data analytics are applied. These elements can come together in a platform approach to real-world evidence generation that Meropol said provides sites with the technology to integrate routinely and intentionally collected data to answer research questions with significantly less site burden than traditional prospective clinical studies (Bourla and Meropol, 2021) (see Figure 4).
Meropol said that many tools to support evidence generation for integrated diagnostics exist and increasing adoption of these tools is occurring. ML tools are available to support automated patient ascertainment. EHR systems have the capability to embed data collection forms for research in routine clinical workflows. And methods exist for the processing of struc-
___________________
31 See https://flatiron.com/resources/case-study-prospective-clinicogenomic and https://clinicaltrials.gov/ct2/show/NCT04180176 (both accessed May 26, 2023).
SOURCES: Neal Meropol presentation March 6, 2023 (Bourla and Meropol, 2021).
tured and unstructured data; automated data transfer from the EHR to data warehouses; and tokenization and linkage of data in the EHR with data from external sources. The next step is to implement these tools on a broader scale to support evidence generation for integrated diagnostics.
Lessons from American Association for Cancer Research Project GENIE to Inform Evidence Generation
Charles Sawyers, founding chairperson of Genomics Evidence Neoplasia Information Exchange (Project GENIE), Howard Hughes Medical Institute investigator and Marie-Josée and Henry R. Kravis Chair in the Human Oncology and Pathogenesis Program at MSK, discussed lessons for evidence generation from his experience with American Association for Cancer Research (AACR’s) Project GENIE.32 The project was launched in 2015, following an agreement among eight cancer centers to aggregate their genomic and clinical data in a public registry. “GENIE was founded on the principle that this knowledge is precompetitive, much like basic science, and should be freely accessible for drug development and drug discovery research,” Sawyers said. A challenge at the start was the need to harmonize data from eight different custom sequencing panels, which Sawyers said was achieved through Sage BioNetworks. OncoTree was selected as the cancer classification platform. He emphasized the importance of stewardship of clinico-genomic registries “to ensure responsible use of the data and protection of patient privacy.”
Project GENIE has 18 contributing cancer centers and will soon be expanding to 22, Sawyers explained. Data are available through cBioPortal,33 and the registry contains nearly 150,000 sequenced tumors (Pugh et al., 2022), with more than 12,000 registered users of GENIE data and more than 800 citations of Project GENIE in publications and abstracts.34
Longitudinal clinical data collection is a focus of Project GENIE, and Sawyers discussed how the GENIE Biopharma Collaborative is curating datasets containing both genomic and clinical data for a range of cancers. The PRISSMM™35 framework is used for curation and incorporates clinical data
___________________
32 See https://www.aacr.org/professionals/research/aacr-project-genie/ (accessed September 5, 2023).
33 See https://www.cbioportal.org/ (accessed February 2, 2024).
34 See https://www.aacr.org/professionals/research/aacr-project-genie/news-updates/ (accessed October 26, 2023).
35 PRISSMM is a cancer data modeling system; it stands for pathology, radiology, imaging, signs and symptoms, tumor markers; and medical oncologist assessments. See https://www.mskcc.org/research-advantage/support/digital-health-projects/prissmm-cancer-data-modeling-system (accessed September 5, 2023).
such as prior treatment history, current therapy, progression-free survival (PFS), and overall survival (Lavery et al., 2022). This work is being funded by the 10 biopharmaceutical partners, and genomic and clinical data for 16,000 patients have been curated so far. Sawyers showed data on checkpoint inhibitor treatment for patients with lung cancer, which demonstrated that determining PFS using PRISSMM is comparable to doing so with the RECIST criteria, which assess disease status based on imaging data.
Sawyers shared several examples of how Project GENIE data are being used, such as to generate external control cohorts for rare disease research (Scharpf et al., 2022; Smyth et al., 2020). In one case, GENIE data were combined with other datasets to create a control cohort that was submitted along with the data from single arm studies of sotarasib36 to support FDA regulatory review and approval of the treatment for a rare type of lung cancer (Scharpf et al., 2022).
Project GENIE data are also being used to explore race and ethnicity differences across cancer genotypes, Sawyers said. One example he mentioned was a study of the frequencies of BRCA237 mutations among Black versus White men with prostate cancer. In another example, Project GENIE data are being used to better understand population-specific miscalling of somatic38 variants, which can inflate somatic mutation estimates among those with non-European ancestry. Studies are also underway to determine genetic ancestry using sequence panels in the registry, compare that with self-reported race, and conduct admixture analysis. Robert Winn, director of the Virginia Commonwealth University Massey Comprehensive Cancer Center, asked about the study of genetic ancestry. For example, triple negative breast cancer is known to disproportionately affect Black women, and Winn referenced a genetic ancestry study that found differences based on Eastern African versus Western African ancestry. Sawyers responded that the intent is to delve into these types of differences with the sequencing panels available. He said that in the absence of whole-genome sequencing of germline DNA, sequencing panels of around 500 genes can provide a remarkable amount of accurate information. Levy
___________________
36 Sotarasib is a KRAS inhibitor approved by FDA as a lung cancer treatment in 2021. The KRAS gene regulates proliferation signals, and mutations can cause unregulated growth, leading to cancer. See https://www.cancer.gov/news-events/cancer-currents-blog/2021/fda-sotorasib-lung-cancer-kras (accessed September 5, 2023).
37 BRCA2, or BReast CAncer gene 2, mutations are associated with increased cancer risk. See https://www.cancer.gov/about-cancer/causes-prevention/genetics/brca-fact-sheet (accessed September 5, 2023).
38 Somatic refers to cells that are not part of the germ line and therefore not passed to future generations. See https://www.genome.gov/genetics-glossary/Somatic-Cells (accessed September 5, 2023).
added that Foundation Medicine has also developed an algorithm based on geographic ancestries. Lennerz noted the lack of consensus on genetic markers based on geographic ancestry data that could be used for validation. Levy noted that even though there are data gaps, genetic geographic ancestry can be useful for increasing diversity among clinical trial populations. Sawyers acknowledged the challenges and added that the human genetics community has achieved “a broader representation of different parts of the world in terms of genome databases” than the cancer community.
A challenge going forward, Sawyers said, is ensuring that data in clinicogenomic registries are generalizable. Project GENIE currently includes four institutions in Europe, and one in Asia will be joining soon. It is also working to increase representation of people of non-European backgrounds in the registry by adding new contributing sites and collaborating with organizations that have genomic data from diverse populations. Sawyers noted that there are barriers to inclusion of persons of non-European ancestry in clinic-genomic registries that need to be overcome, such as meeting the infrastructure requirements for centers to contribute to registries (e.g., ability to deliver the type and quantity of data needed, full informed consent processes, and appropriate data stewardship) and building the trust needed to increase participation of diverse communities in research. Levy added that there are specific challenges for including data from non-U.S. institutions, including different approaches to regulation and ensuring patient privacy. The diversity of the data in Project GENIE is based on the diversity of the patient populations of the contributing institutions, and institutions that serve more diverse populations are also those that often do not yet have the infrastructure needed to participate. Sawyers said that Project GENIE generally does not have the resources to support this infrastructure but can sometimes assist with start-up costs. He also noted that some centers that do have the infrastructure but prefer not to participate.
Mark Stewart, vice president of science policy at Friends of Cancer Research, raised the issue of variability in the performance of different diagnostic assays with the same intended use and how that might affect harmonization and interpretability of data from different institutions. Levy responded that Project GENIE datasets include data from different assays, and the ability to ask a particular question will be limited by whether a gene was included in particular assays. Another issue, discussed by Trentadue, is the need for a clear ontology for diagnostic procedures. Levy said, for example, that testing menus from different institutions might call the same diagnostic test “a right-sided mammogram, a right-side diagnostic mammogram, right-sided mammo, or R mammo.”
The Role of Equitable Access in the Diversity of Registry Data
Many workshop speakers discussed the role of patient access in ensuring the diversity of the populations in clinic-genomic registries. Meropol said a key impediment to representation of diverse patient populations in clinical trials is whether individuals actually have access to a clinical trial, including awareness that it exists and the ability to participate. He advocated for increasing clinical research sites in community settings of cancer care. This would improve patient access to clinical trials, and the data from these trials will better reflect the diversity of the people receiving cancer care in the community.
Hirsch added that another element of improving access is having a user experience team that understands nuanced differences across geographies, among those with different socioeconomic status, and other community factors to improve engagement with diverse participants.
Winn highlighted the need to engage not only institutions that have high tech but also those that have “high touch.” Specifically, he suggested leveraging existing national infrastructures such as the NCI Community Oncology Research Program (NCORP)39 and Federally Qualified Health Centers.40 Hirsch pointed out that many oncology clinical trials are commercially sponsored and often conducted by large cancer networks that have a single EHR system and single research infrastructure. He suggested that institutions participating in NCORP might be less attractive from a study sponsor’s perspective and encouraged discussion of incentives to facilitate participation.
DESIGN AND USE OF INTEGRATED DIAGNOSTICS
To realize the potential of integrated diagnostics for precision oncology care, many speakers noted that institutions will need to consider implementation of appropriate tools and technologies to ensure clinician adoption. A range of design and use challenges were discussed, and many speakers highlighted potential solutions to promote acceptance and uptake of integrated diagnostics.
Ethical AI by Design
As the data science continuum moves from traditional statistical analyses toward ML, models become increasingly more accurate, but interpretability decreases, said My Thai, associate director of the Nelms Institute for the Connected World and the Research Foundation Professor of Computer and
___________________
39 See https://ncorp.cancer.gov/ (accessed September 5, 2023).
40 See https://www.fqhc.org/what-is-an-fqhc/ (accessed January 27, 2024).
Informational Science and Engineering at the University of Florida. AI models in diagnostics are increasingly computationally complex, with an emphasis on prediction speed and quality but with less transparency. These models can lead to unintended consequences, including shortcut learning (unintended learning from the training materials), which may contribute to unexpected failure of algorithms in broader clinical practice.
Thai said another concern is the potential for bias. Training datasets are collected and annotated by humans, making them subject to human biases. Encoded biases in the dataset are amplified during training, leading to biased outputs. “In some cases, the output data becomes the training data for the next model,” she said, leading to an ongoing cycle of bias amplification.
Thai noted the patient privacy and data security concerns associated with ML and described several types of adversarial attacks that can occur in AI design. The training phase can have “poisoning” of the training data with data intended to alter model performance. In the inference phase, when the predictive model is deployed, adversarial attacks can include “membership inference,” an attempt to reverse engineer the model to predict what data were included in the training set; “model extraction,” in which the outcome is used to steal the functionality of the model; and “adversarial examples,” which are noisy data added to the input with the intent to cause the model to misclassify outcomes.
To address these concerns, Thai listed six key elements to design ethical AI models: they should be transparent and understandable, inclusive, responsible, impartial and unbiased, trustworthy and reliable, and able to ensure data security and user privacy. Thai also emphasized the need to ensure that training datasets are appropriately balanced with respect to population diversity and assess whether the model, as designed, applies equally to everyone—a key aspect of the “inclusion” element of ethical AI.
Thai also said that designing ethical AI is not just the domain of computer science but requires multidisciplinary collaboration. This approach focuses on data security and patient privacy, explainable AI, and fairness in the development of an AI model with high predictive accuracy (Phan et al., 2019, 2020). She noted that there is little value to developing a secure, fair, explainable model that is inaccurate and explained that these elements are overlapping. For example, data security and patient privacy are important to explainable AI (e.g., blocking new avenues of adversarial attacks) and fairness (e.g., minimizing risk of exposure of sensitive information, such as age, sex, and race). It is also important to be alert to the creation of new problems and address them in the development process. For example, adversaries can query the model to receive an explanation of how the AI derived the prediction and then exploit that information to devise an attack. The challenge, Thai said, is how to provide useful explanations to users without enabling adversarial attacks (Nguyen et al., 2022).
A Diagnostic Management Team Approach to Reducing Diagnostic Errors
The volume of knowledge about disease has increased vastly over the past 70 years, and it is simply not possible for any clinician to know everything about a specific area of disease, said Michael Laposata, professor and chair of the Department of Pathology at the University of Texas Medical Branch (UTMB). Diagnostics such as imaging studies and biopsies are routinely interpreted by highly trained, specialized diagnostic physicians—radiologists and anatomic pathologists, respectively. However, for laboratory testing, Laposata said that a patient’s clinician has to “select all the correct tests and interpret the results with little or no assistance.” This can lead to diagnostic errors, which are a significant contributor to patient harms (NASEM, 2015).
Laposata discussed the challenges clinicians face when interpreting coagulation test results as a case example of the need to optimize diagnostic medicine. Laposata has been advancing the concept of the diagnostic management team (DMT), which includes “experts who can recommend when to use which diagnostic tests and then interpret diagnostic testing results in highly specific disease categories,” he explained. The goals of a DMT are to “shorten the time to diagnosis, increase the accuracy of diagnosis, and optimize the utilization of laboratory tests.” A barrier to this approach, however, is that payers often do not cover the time spent by expert physicians consulting on diagnostic test ordering and interpretation. To address this, Laposata and others worked to develop and implement billing codes for the time laboratory medicine experts spend advising on test selection and interpreting results. Through the DMT approach, physicians with expertise in clinical pathology, genetics, radiology, anatomic pathology, and history and physical exam are able to integrate all relevant diagnostic and clinical data to provide the ordering clinician with a diagnostic testing strategy and provide an interpretation of the test results.
Laposata said the DMT approach has been implemented at UTMB for testing associated with coagulation, toxicology, autoimmunity, complex transfusions, pharmacogenomics, anemia, liver disease, COVID-19 infection, and others. Implementing DMT support for coagulation testing at UTMB in 2014, for example, has resulted in a steady decline in hospital length of stay for patients with coagulation disorders and was correlated with reaching a faster diagnostic conclusion that led to a path for care, said Laposata. In another example, he said DMT interpretation of testing for COVID-19 at UTMB was associated with decreased length of stay compared to other U.S. academic medical centers and a lower rate of early deaths (unpublished results). Laposata explained that providing an actionable interpretation of COVID-19 test results to patients through the EHR patient portal may have contributed
to those improved outcomes. He added that the work of the DMTs has demonstrated significant value for the hospital system.
Having implemented a DMT approach at UTMB and other institutions, Laposata is working on efforts to create a national DMT, in which a patient’s clinician would reach out to a local diagnostic physician with questions, who would provide answers based on their knowledge and consult with national-level DMTs for further assistance as needed. The local physician would provide the information from the national DMT to the patient’s clinician, who can then act on the experts’ recommendations. If done well, Laposata said, a national DMT model could facilitate rapid and accurate diagnoses that improve patient outcomes.
Multimodal Data Integration for Research and Clinical Practice
Sohrab Shah of MSK explained that computational research can facilitate insights from real-world data, multimodal data integration, cohort derivation and comparison, and patient stratification and predictive models (Boehm et al., 2022a, 2022b). MSK has long invested in developing platforms for cancer data science, Shah said, including the following:
- cBioPortal for cancer genomics datasets41
- MSK MIND (Multimodal Integration of Data)42
- OncoKB (a precision oncology knowledge base)43
- Warren Alpert Center for Digital and Computational Pathology44
Shah said that the understanding of cancer biology is informed by experimental data on cancer progression and drug resistance, the tumor micro-environment, cellular phenotypes, interactions among tumor cells and immune cells, and multiomics data. He said that applying computational oncology approaches to these data can facilitate the development of new cancer treatments and integrated diagnostics.
As proof of principle for clinical application, Shah described an example of multimodal real-world data integration to predict response to immunotherapy in patients with non-small-cell lung cancer. The lung tumor experts at MSK were facing challenges predicting who would respond to an immune
___________________
41 See https://docs.cbioportal.org/about-us/ (accessed September 5, 2023).
42 See https://www.mskcc.org/research-programs/msk-mind-multi-modal-integration-data (accessed September 5, 2023).
43 See https://www.oncokb.org/ (accessed January 25, 2024).
44 See https://www.mskcc.org/departments/pathology-laboratory-medicine/warren-alpert-center-digital-and-computational-pathology (accessed September, 5, 2023).
checkpoint blockade therapy. Shah and colleagues developed a computational workflow and ML approach to extract and integrate genomic, histology, and radiology data to provide a prediction of a patient’s response to therapy (Vanguri et al., 2022). The development process was labor intensive, said Shah; a thoracic pathologist reviewed and labeled all tumor regions in digitized immunohistochemistry images. “We then started to compute the topological features of the staining properties within each of the tumor regions that she labeled,” he explained. This took approximately 1.5 years, with a comparable process for the radiology images. Shah said that the integrated multimodal data approach provided superior predictive performance compared to several predictive approaches that involved only one modality (Boehm et al., 2022a; Vanguri et al., 2022; Vázquez-García et al., 2022).
Shah also described a study integrating multiomic data to elucidate the natural history of disease for ovarian cancers, particularly the relationship between genomic DNA damage and the immune response (Vázquez-García et al., 2022). This was also a labor-intensive, multidisciplinary process involving surgical collection of tumor tissue, whole genome sequencing, single-cell RNA sequencing, digital pathology, and phenotypic profiling of immune cells.
“Multimodal data integration is a very powerful approach for both real-world and experimental data interpretation,” Shah summarized, and he shared several lessons. It requires both clinical expertise and computational rigor, and he emphasized the need for multidisciplinary collaboration. He suggested that large-scale retrospective multimodal studies are needed to inform integrated diagnostics but also noted that obtaining data for these can be challenging. He explained that multimodal data integration at scale requires data engineering, because diverse types of data need to be drawn from disparate sources into a centralized repository that can be queried and used for ML models. Reproducibility in ML is essential, but Shah noted that can be difficult and different institutions need to validate the models. Finally, Shah noted that the field faces a talent “bottleneck,” and more data scientists need to be recruited to the field of cancer research and care.
Implementing Integrated Diagnostics into Precision Oncology Care
David Dorr, chief research information officer and vice chair of Medical Informatics and Clinical Epidemiology at Oregon Health & Science University (OHSU) School of Medicine, defined health informatics as the science of the use of data, information, and knowledge to improve health.45 Dorr discussed ways in which the multidisciplinary field of health informatics
___________________
45 See https://amia.org/about-amia/why-informatics/informatics-research-and-practice (accessed January 25, 2024).
seeks to understand how users interact with data and to mitigate cognitive and data biases in the use of these data. From a cognitive perspective, several key elements determine whether and how people act on computer-aided decision support recommendations for patient care, and Dorr referenced the affect-integration-motivation and attention-context-translation framework (Nahum-Shani et al., 2022).46 Key issues that affect clinician uptake of findings from digital interventions such as integrated diagnostics include trust, understanding, timing, options, prioritization, adjudication and annotation, and actionability. Given the many things that compete for a clinician’s time and attention, Dorr emphasized that a synthesis of findings from an integrated diagnostics tool needs to make clear what the most important information is, why it is important for their patient at that time, and what actions can be taken.
Dorr described a study comparing algorithmic risk scoring to clinical intuition in predicting which patients were most likely to be hospitalized within the coming year (Dorr et al., 2021). The study found that algorithms were more accurate than clinical intuition for risk prediction and that clinician adjudication of algorithmic risk scores improved performance further, which he said was associated with confidence and trust in the risk scoring process.
Dorr pointed out that most algorithms rely on EHRs and other real-world data sources, which may contain errors or conflicting information. For example, diagnoses appear in multiple EHR locations and can be inaccurate or missing, impacting predictions (Martin et al., 2017). Dorr added that missing data are not necessarily random and can be associated with inequities.
Data may also contain biases that reflect societal biases, Dorr continued, and algorithms can perpetuate biases depending on how they are designed. Dorr cited an example in which an algorithm predicted that Black and White patients have the same level of risk for future health care use (Obermeyer et al., 2019). However, Black patients experienced more health problems that would likely necessitate higher future health care utilization. In this case, the underlying bias was inequitable access to health care: the algorithm predicted future risk based on spending data, which was more reflective of a patient’s access to care and not on their health status.
Integrating advanced algorithms into the clinical workflow also presents challenges. Dorr discussed the role of implementation science in promoting the uptake of integrated diagnostics and preventing unintended consequences. He mentioned the Consolidated Framework for Implementation Research as an example of one effective approach (Damschroder et al., 2022).
___________________
46 The affect-integration-motivation and attention-context-translation framework is intended to “provide recommendations for designing strategies to promote engagement in digital interventions and highlight directions for future research” (Nahum-Shani et al., 2022).
At OHSU, the Care Management Plus team is focused on “improv[ing] systems and outcomes for vulnerable populations through research, technology, and collaboration,” Dorr explained. Researchers are working to identify vulnerable populations through risk stratification and tailor care to meet their needs. OHSU is “eager to implement advanced algorithms,” he said, “but also cautious” given the pros, cons, and challenges for deploying deep learning methods in clinical care (Egger et al., 2022; NASEM, 2022). In closing, Dorr shared the code of conduct for implementation of AI at OHSU:
- Demonstrated clinical utility;
- A clear and evidence-based risk–benefit calculation;
- Consideration of the ethics of the model, especially when vulnerable patient populations are involved;
- Local validation and evaluation;
- An implementation plan that includes monitoring for benefits and harms;
- A training and support plan to help all persons at OHSU, including patients, understand and use the tools effectively and safely; and
- Appropriate integration with the EHR to support clinician needs.
Taking a Holistic Approach to Health Care
“From my first day as a nurse, I knew that the EHR system had to change,” said C. J. Robison, now a health innovation scientist with the Oracle Health Global Business Unit. Robison posited that the EHR should be conceptualized less as a digitized patient record and more as a system of tools that “work together seamlessly to optimize … care delivery for the patient.” For patients with cancer and their caregivers, a holistic approach can make it easier for them to navigate through a complicated care system. Robison said this means not simply returning results to the patient but helping them understand what to do with that information.
For clinicians, a holistic approach makes it “easy to do the right thing,” Robison said. All health care staff are increasingly pressed for time, and systems need to make the “right” options the easiest, she said. From an operations perspective, Robison said that a holistic approach provides opportunities to improve patient and staff experiences (e.g., optimizing staffing schedules based on infusion time).
Robison highlighted three areas to better align EHR functionality with clinician needs:
- Data visualization platforms to help the clinician quickly understand their patient’s situation. Clinicians are spending their limited time
- clicking through EHR pages to hunt for images or laboratory results, and they are eager for an easier way to get the most important information about their patient, Robison said, including correlation with data elements from longitudinal records.
- ML for testing and workflow to help clinicians understand what to do next. In a survey of oncologists, 79 percent discussed genetic testing with their patients, Robison said, but only 12 percent reported that at least one of their patients had undergone genetic testing (Culver et al., 2001). ML can nudge clinicians toward potentially relevant testing or treatment options based on patient results, new research about a treatment, or other information.
- A multilevel modeling system to help avoid silos and create “a holistic system in which all of our models work together, generat[ing] a seamless ability for the clinician to make the right decision for their patient at any moment.”
Robison said that a holistic approach to research drives progress by incorporating data from a range of sources (e.g., clinical, patient-reported outcomes, remote patient monitoring) but also noted the challenges of building structures for big data that can help facilitate patient care today and retrieval and use in the future, as tools and technologies advance. When working to create integrated systems, Robison concluded, it is important to “deeply understand the needs of the clinician” and structure systems that eliminate silos and “work with the cognitive flows that happen at that point of care.”
Trust and Acceptance of AI in Clinical Settings
Despite the growing evidence supporting the use of AI in precision oncology to streamline the workflow, accelerate diagnosis, and improve the quality of patient care, many clinicians still hesitate to use it in practice, said Avishek Choudhury, assistant professor of Industrial and Management Systems Engineering at West Virginia University’s Benjamin M. Statler College of Engineering and Mineral Resources.
Choudhury noted that a key factor in adoption is trust in the technology. A clinician who trusts AI is willing to make or change their decision based on its recommendation, Choudhury said. He added that it is important to distinguish between trust and confirmation bias (when the user only trusts AI when it confirms their beliefs) because the latter case sacrifices the potential for AI to fill gaps in human performance. He said protocols or guidelines are needed for situations with discordance between the algorithm and clinician conclusion. These would guide the clinician in reconsidering or changing their diagnosis or updating the algorithm accordingly.
Choudhury also described a parallel integration approach under investigation in which the clinician is not aware that an algorithm is running simultaneously. The final decisions are compared, and the AI learns from the clinician decision and patient outcomes. Upon completion, the clinician is notified and thanked for helping to train the algorithm. He suggested that this approach helps to change perceptions of AI and gathers use case data for full implementation in the future.
The willingness to trust involves both initial trust, which is based on assumptions and perceptions, and trust evolution, which stems from experience and consequences, Choudhury explained (Choudhury and Elkefi, 2022). Accordingly, a clinician’s willingness to try AI when presented with the option depends on their initial trust. A challenge is that AI is often perceived negatively (e.g., as a threat, because it may be “better” or more efficient than the clinician or disregards their expertise), and Choudhury emphasized the importance of improving health professionals’ understanding and perception of AI. Given initial uptake in routine clinical care, feedback on patient outcomes and AI performance (e.g., ethics, privacy, generalizability) affects the evolution of trust. Approaches to optimizing trust in AI include increasing transparency, ensuring robustness, and encouraging fairness (Asan et al., 2020). Choudhury emphasized that the goal is optimizing and not maximizing trust and cautioned that maximized trust is blind trust, which can lead to biased outputs that adversely impact health outcomes.
Choudhury described an example of a hospital that used an AI-based application to predict how many units of blood a patient is likely to use in order to reduce unnecessary blood transfusion. Although it had been shown that the algorithm performed better than the clinicians at this task, clinicians only looked at the AI recommendation 46 percent of the time (Choudhury et al., 2022). He added that junior clinicians faced with an AI recommendation that differs from their own conclusion tend to confer with and follow the advice of the attending physician instead.
Accountability is another element that affects clinician trust in the system and AI uptake. If a clinician relies on an AI recommendation and the patient experiences harm, it can be challenging to establish the source of the harm and raises questions about where accountability lies (Habli et al., 2020). In a recent survey, clinicians expressed concerns about accountability, for example, that using AI could affect their career or lead to losing their medical license (Choudhury and Asan, 2022).
Trust in the system is also affected by clinician perceptions of how AI affects their workload, Choudhury said. Some clinicians surveyed felt that AI added to their workload (e.g., additional trainings, another module in the EHR to navigate, time needed to explain to patients the role of AI in treat-
ment decisions). He offered three strategies to enhance trust in the technology and improve acceptance and adoption (Choudhury, 2022):
- Establish shared or distributed accountability based on risk and intended use.
- Take human factors into consideration (e.g., trust, perceptions, workload).
- Implement need-based integration to help end users to understand how AI is a tool that can help them improve effectiveness and efficiency.
Risk Communication and Decision Support
Patient perception and understanding of health risks vary, said Mary Politi, professor in the Department of Surgery at Washington University School of Medicine. Clinician abilities to explain clinical uncertainty to their patients and manage patient care in the context of uncertainty can also vary. She referenced an interview-based study that identified four categories of strategies clinicians use for uncertainty management, focused on ignorance, uncertainty, response, and relationships (Han et al., 2021). The first two approaches seek to reduce the uncertainty, and the last two seek to cope with its effects. Politi centered her remarks on the relationship-focused tactic of sharing information about clinical uncertainty with patients. She quoted a clinician in the study who discussed the science versus the art of medicine and the need to embrace the uncertainty and share information with the patient to help them make personalized choices about their care.
Politi described the three-talk model of shared decision making: a “team talk” to establish the clinician–patient partnership in treatment decisions; an “option talk” using risk communication strategies to inform patients about their care options; and a “decision talk” to incorporate patient needs and preferences in the care decision (Elwyn et al., 2017). The option talk includes risk prediction models incorporating the results of diagnostic testing. Politi summarized some central tactics of communicating risks, benefits, and uncertainty to patients during the option talk:
- Using numbers. Avoid imprecise terms, such as “low,” “moderate,” or “high,” when discussing risk. “Without numbers, people will underestimate their risk and overestimate their benefit,” Politi said.
- Using frequencies and keeping the denominator consistent. Make it easy for patients to make comparisons (e.g., do not present the likelihood of one outcome as 1 in 10 and another as 1 in 12).
- Using visuals. Pictures can help to convey risk information more clearly. Many risk communication studies have suggested the use of icon arrays.
- Providing a reference group and time period, and using absolute (vs. relative) risk. When discussing population-level risk data, patients often need help to understand how the population and time period studied relate to their situation; it is clearer to be told the absolute risk (e.g., that a treatment reduces their risk from 2 in 1,000 to 1 in 1,000) than the relative risk (e.g., that it reduces their risk by 50 percent), Politi said.
Politi shared several examples of clinical decision support tools that employ these risk communication principles. One tool, to predict the risk of sentinel node metastasis in melanoma, is designed for the patient encounter. The report with the results helps to guide discussion of the patient’s personal risk and informs shared decision making about the need for central node biopsy.47
The second example described by Politi was BREASTChoice, a tool designed for patients who have had a mastectomy to help them understand their personal risks in breast reconstruction surgery (Foraker et al., 2023; Lee et al., 2022; Politi et al., 2020).48 Politi mentioned there can be risks for serious post-surgical complications of reconstruction, depending on a patient’s risk factors. The tool uses data from the patient’s EHR to automatically populate the risk prediction model to provide a comparison to a reference group of individuals who have no risk factors (including a visual aid). A summary of the assessment is also sent to the clinician to discuss during a patient visit. Politi said that this approach of interfacing the algorithm with the EHR is still evolving and preferences for how to enter patient data vary. “Most of the literature suggests that people don’t want to enter this data on their own,” she said. However, she noted that the challenges with automatically retrieving the necessary data from the EHR (e.g., data missing from fields, nonstandardized or free-text elements), and lack of interoperability of EHR systems would present challenges for scalability.
Politi provided a nononcology example to demonstrate the value of clinically relevant reference groups for patients to compare out-of-range laboratory results in the patient portal: a patient with diabetes sees their hemoglobin A1C results relative to the goal range for a people with Type 2 diabetes (not the standard reference range).49 Studies have found that this approach helps patients to better interpret their out-of-range results and take action accordingly (Scherer et al., 2018).
___________________
47 This tool is publicly available from the Melanoma Institute Australia at https://www.melanomarisk.org.au/SNLForm (accessed May 26, 2023).
48 See https://breastchoice.wustl.edu/ (accessed September 5, 2023).
49 The A1C test is used to measure a patient’s average blood sugar levels and can be used to diagnose or monitor prediabetes and diabetes. See https://www.cdc.gov/diabetes/managing/managing-blood-sugar/a1c.html (accessed September 5, 2023).
A study by Politi and colleagues found that clinicians have different preferences for risk communication tools, such as conversation aids (e.g., paper or digital; before, during, or after the visit), and that choice often depends on the context of use (Politi et al., 2015). These tools can provide clear language for clinicians to use in difficult discussions about the potential for serious risks of an intervention (Hasak et al., 2017).
Gwen Darien, executive vice president of patient advocacy and engagement at the National Patient Advocate Foundation and a three-time cancer survivor, agreed that clear risk communication, including discussion of absolute risk, is essential for patients making care decisions. She shared a personal story of the negative impact of not being told about potential adverse effects of a treatment. When she asked why, her oncologist told her he did not want to cause her any worry. Darien stressed that “patients can understand so much more than they are given credit for” and want to be equipped with the available information to make informed decisions about their care.
Reproducibility of ML Models for Integrated Diagnostics
Salto-Tellez pointed out that an AI algorithm developed by one institution, using training data from it, often does not perform as expected or required at a different institution. He asked what factors might be contributing to this lack of reproducibility for integrated diagnostics (e.g., quality of the algorithm or the clinical information). Shah said that overfitting, or the inability of a model to generalize due to limitations of its training, is challenging to address. He shared that the reviewers of his manuscript on the algorithm to predict response to an immune checkpoint blockade therapy asked whether it could be validated in an external cohort, and efforts to find an appropriate external dataset were unsuccessful. Shah said that the data behind the predictive algorithm have been publicly released and can be used by others to train their models (Vanguri et al., 2022), and he advocated for the concept of open data. Although some institutions are releasing genomic sequencing data through collaborations (e.g., Project GENIE, discussed by Sawyers), he explained that researchers often cannot access clinical sequencing data even from within their own institution. “This field will not move forward until we embrace open data,” he said. Elenitoba-Johnson agreed that it is essential to make algorithms available for testing by others.
Dorr said that federated datasets can be used to study the reproducibility of an algorithm in other settings. The Observational Health Data Sciences and Informatics (OHDSI)50 federated dataset, for example, includes data from
___________________
50 See https://www.ohdsi.org/ (accessed September 5, 2023).
810 million patients. Dorr explained that variation in algorithm performance is to be expected and provides opportunities for learning. Shah agreed that federated learning could be an option for algorithm validation and reproducibility studies, in which the algorithm is trained on datasets at multiple institutions and then the models are integrated. Shah cited a recent report of federated learning for predicting response to a treatment for breast cancer (Ogier du Terrail et al., 2023).
Regina Barzilay, MacArthur Fellow and School of Engineering Distinguished Professor for AI and Health in the Department of Electrical Engineering and Computer Science at Massachusetts Institute of Technology’s Computer Science & Artificial Intelligence Laboratory, suggested that another reason for lack of reproducibility is that algorithms in health care are not as robust as those in other contexts. Existing software is often adapted for use with medical data without “the same degree of mathematical sophistication and engineering.” As an example of a robust algorithm, she noted that “it is really hard to find a place where your face will not be recognized” by an iPhone’s facial recognition software.
Barzilay also noted the role for publishing standards that foster reproducibility and that some journals expect demonstration of reproducibility across institutions as a requirement for publication.
Overcoming Design and Use Challenges
Many speakers suggested potential opportunities for overcoming the challenges related to designing and using integrated diagnostics in precision cancer care. Robison, referencing the challenge of EHR organization of information by type (e.g., laboratory data are in one location, images are in another) suggested that the data visualization platforms should be contextually aware, presenting the data that are needed in that moment for the decision. Shah agreed that organization should be patient centric rather than by data type. He recalled Comaniciu’s discussion of digital twinning and said MSK is exploring the concept of a user interface built around a digital patient, but implementation challenges remain, such as how data are weighted and selected for context-relevant inclusion in the presentation. Dorr said that some type of “annotation or adjudication of what is most important” is needed to determine “what data should be prioritized at what time.”
Dorr highlighted the need for research on understandability and actionability of information. How do clinicians interact with the volumes of data related to their patients, especially those with complex conditions, and what is it that they actually look at? He said health care algorithms need to be adapted to include data elements that clinicians use. “There is real harm that’s done when data are missed,” he cautioned. He suggested looking to the field of
industrial engineering to understand how to insert information into different workflows.
Politi observed that graphics on decision aids can be very complicated and include animations and other features. While visually appealing, she explained that some more complex presentations can be distracting for the patient receiving the information. She advocated for clear, simple diagrams that can be personalized as needed.
Choudhury suggested developing AI algorithms to assess whether a patient is at risk for severe depression or suicidal ideation when a clinician is delivering a diagnosis. This could inform how care is delivered, including provision of additional mental health support.
REGULATORY OVERSIGHT AND INSURANCE COVERAGE OF INTEGRATED DIAGNOSTICS
Several speakers discussed the importance of achieving regulatory clearance or approval for clinical use and establishing coverage and reimbursement mechanisms for the adoption of integrated diagnostics.
Pathways for Regulatory Review of Integrated Diagnostics
Reena Philip, associate director for Biomarkers and Precision Oncology at the FDA Oncology Center of Excellence, provided an overview of the current FDA review framework for IVDs, radiological devices, and AI-based digital pathology and radiological devices.
Philip explained that the regulatory pathway for all medical devices begins with the submission of an application for premarket review of safety and effectiveness by FDA’s Center for Devices and Radiological Health (CDRH). Applications are processed differently according to the device’s risk classification (Box 3). Upon clearance or approval, the device may enter the market for clinical use. Safety and effectiveness monitoring continues through postmarket surveillance activities including required reporting of serious and adverse events associated with device use. The FDA Total Product Life Cycle database integrates premarket and postmarket data about medical devices, including about adverse events and recalls.51
___________________
51 See https://www.fda.gov/about-fda/cdrh-transparency/cdrh-transparency-total-product-life-cycle-tplc (accessed May 26, 2023).
BOX 3
FDA Risk Classification–Based Review Pathways
Class I
Devices with a low risk of illness or injury. Most are exempt from FDA review.
Class II
Devices with a moderate risk of illness or injury. Some are exempt from review. Most will require submission of a 510(k) premarket notification to receive FDA clearance to market.
Class III
Devices that present a high risk of illness or injury. If a predicate device exists, the new device may be able to a submit a 510(k) premarket notification to receive FDA clearance to market. If no predicate device exists, it must undergo the full FDA premarket approval process, which includes submission of valid scientific evidence of safety and efficacy.
SOURCES: Reena Philip presentation, March 6, 2023. See also https://www.fda.gov/patients/device-development-process/step-3-pathway-approval (accessed May 26, 2023).
In Vitro Diagnostics
Philip said FDA defines IVD devices to include “those reagents, instruments, and systems intended for use in the diagnosis of disease or other conditions, including a determination of the state of health, in order to cure, mitigate, treat, or prevent disease or its sequelae” (21 CFR 809.3). She provided examples of IVDs in each class: a test for serum prealbumin or a nucleic acid extraction kit would be Class I; an autosomal recessive carrier screening gene mutation detection system would be Class II exempt, but a next-generation sequencing-based tumor profiling test would be Class II requiring 510(k) clearance; and a colon cancer screening test or a companion diagnostic test52 would be Class III requiring premarket approval.
___________________
52 A companion diagnostic, usually an IVD, is a medical device that provides necessary information on how to use a drug or biologic product safely and effectively. See https://www.fda.gov/medical-devices/in-vitro-diagnostics/companion-diagnostics (accessed January 28, 2024).
Radiological Devices
Philip also gave examples of radiological devices in each risk class: ultrasound gel or gloves are Class I and most are exempt; MRI scanners or catheters would be Class II requiring 510(k) clearance; and radioactive microspheres or stents would be Class III requiring premarket approval. She pointed out that FDA is charged with regulating the manufacturers of the equipment and the equipment itself, while the use of these devices is regulated by other Federal agencies, including the Nuclear Regulatory Commission, the Occupational Safety and Health Administration, and the Environmental Protection Agency, as well as state and local agencies. The quality of care provided by the health care organizations where the devices are used is overseen by accrediting bodies, including the Centers for Medicare & Medicaid Services (CMS), American College of Radiology, and The Joint Commission.
AI-Based Digital Pathology and Radiological Devices
FDA defines AI as a device or product that can imitate intelligent behavior or mimics human learning and reasoning. AI includes ML, neural networks, and natural language processing. Philip said that terms used to describe AI include computer-aided detection/diagnosis, statistical learning, deep learning, or smart algorithms. Philip said an example of an AI-based device would be “an imaging system that uses algorithms to provide diagnostic information for malignant melanoma or skin cancer in patients.”53
Regulation of AI applications also follows the risk-based approach. In digital pathology, Philip explained that the intended use is considered, including whether it is to be added to or replace the standard of care. She noted that most such devices are currently image based and that “differences in AI device performance based on differences in digital images should be assessed.” AI applications in radiology spans the imaging continuum: from acquisition to reconstruction, filtering, denoising, interpretation, and reporting. Philip noted that a listing of the AI and ML devices FDA has reviewed is available on the agency’s website.54
Providing an example of an FDA-cleared AI-based device, Philip pointed to an AI-based software for the detection of areas that are suspicious for prostate cancer developed by MSK and Paige.AI.55 This Class II device is a
___________________
53 See https://www.fda.gov/medical-devices/digital-health-center-excellence/digital-health-terms (accessed May 26, 2023).
54 See https://www.fda.gov/medical-devices/software-medical-device-samd/artificial-intelligence-and-machine-learning-aiml-enabled-medical-devices (accessed May 26, 2023).
55 See https://paige.ai/ (accessed September 5, 2023).
locked algorithm, with its performance dependent on the training dataset. She said that product sponsors needed to ensure sufficient data to support the intended uses and that the intended patient population is represented in the validation dataset.
In response to a question, Philip said that, to her knowledge, all AI-based devices reviewed by FDA thus far have been locked algorithms. In reviewing adaptive algorithms, she said it would be necessary for FDA to understand up front how performance of one deemed safe and effective could be monitored after clearance or approval to ensure ongoing safety and effectiveness in the marketplace. She added that research is needed to determine how to perform this monitoring.
Opportunities and Challenges
AI-based medical devices can use real-world data to continuously learn and improve and have the potential to “transform the delivery of health care,” Philip said. But challenges remain, including creating the “large, high-quality, well-curated datasets” needed to train models; explaining AI algorithm decision making (i.e., “black box”56 approaches); identifying and eliminating biases in models; and ensuring transparency for users.
Philip highlighted several FDA activities to address these challenges. The FDA–NIH Joint Leadership Council Working Group on Next Generation Sequencing and Radiomics explored the development of reference materials to support the validation of next-generation sequencing tests and the use of AI and ML to interpret next-generation sequencing and radiomics data. She referred participants to the highlights of a workshop in September 2021.57 Related initiatives with AI-focused activities include the following:
- Medical Device Innovation Consortium Somatic Reference Sample Initiative,58
- Digital Health Center of Excellence-AI and Machine Learning Software as a Medical Device Action Plan,59
___________________
56 See https://www.scientificamerican.com/article/why-we-need-to-see-inside-ais-black-box/ (accessed November 1, 2023).
57 See https://dctd.cancer.gov/NewsEvents/20211122_NCI_Hosts_FDA_NIH_Workshop.htm (accessed May 26, 2023).
58 See https://mdic.org/wp-content/uploads/2022/10/MDIC-Initiative-SRS_CS_FINAL-Web.pdf (accessed January 25, 2024).
59 See https://www.fda.gov/news-events/press-announcements/fda-releases-artificial-intelligencemachine-learning-action-plan (accessed January 25, 2024).
- The CDRH 2022–2025 Strategic Priority to Advance Health Care Equity,60 and
- NCI Cancer Research Data Commons Repositories.61
Finally, Philip called attention to precisionFDA,62 “a secure, collaborative, high-performance computing platform that builds a community of experts around the analysis of biological datasets in order to advance precision medicine.” Crowdsourcing challenges hosted by precisionFDA include advancing regulatory standards for the use of real-world data and AI.63
Coverage Considerations for Integrated Diagnostics
Jennifer Malin, senior vice president and chief medical officer at Optum Health Solutions, reviewed four key elements for establishing coverage and reimbursement of a health care technology in a fee-for-service environment:
- Benefits. The first question is whether the diagnostic technology is included in a covered category of benefits. Although standard benefits generally include diagnostic testing and pathology imaging, diagnostics that incorporate AI might not be covered, and creating a new category for certain integrated diagnostics might be needed.
- Coding. Billing a fee for a service requires a Current Procedural Terminology (CPT) code. If no code can be used to bill for an integrated diagnostic, a process64 needs to be followed to assign a new code.
- Clinical coverage. Having a CPT code does not guarantee reimbursement. A favorable clinical coverage determination generally requires a high level of evidence of clinical utility, and population-based screening tests for risk prediction “require a very high level of evidence,” Malin pointed out, adding that Medicare and private payers generally follow the recommendations of the U.S. Preventive Services Task Force. Although most private payers will mirror CMS coverage determinations, she noted that “reimbursement for genetic testing varies dramatically across state Medicaid plans.”
___________________
60 See https://www.fda.gov/media/171713/download (accessed January 25, 2024).
61 See https://datascience.cancer.gov/data-commons/repositories (accessed January 25, 2024).
62 See https://precision.fda.gov/ (accessed March 6, 2024).
63 See https://precision.fda.gov (accessed May 26, 2023).
64 See https://www.ama-assn.org/about/cpt-editorial-panel/cpt-code-process (accessed January 28, 2024).
- Reimbursement. Once a determination had been made to cover a service, a reimbursement rate is set.
Malin discussed genetic testing as an example of demonstrating clinical utility for a coverage determination. “To have clinical utility, a genetic test must lead to an action that has been shown to improve patient outcomes,” she said. For example, the test might identify a mutation that could guide the choice of a targeted therapy, thereby improving patient outcomes. It might also lead to a diagnosis or prediction that results in a change in clinical management that produces an improved patient outcome. For integrated diagnostics, overall survival is a key outcome measure; however, intermediate outcomes are also important (e.g., timeliness of initiation of therapy, reduction in the amount of testing needed). She emphasized the need to clearly define the improvement in outcome that integrated diagnostics provide and added that demonstrating population-level improvements in health outcomes can take decades.
As health care financing strategies evolve from fee-for-service to value-based payments, health systems have increasing flexibility in structuring how care is delivered, and Malin said this could provide greater opportunities for incorporating integrated diagnostic approaches. Value is defined as the outcomes relative to cost (Porter, 2010), and a value-based care model provides reimbursement based on indicators of value (e.g., outcomes, efficiency, quality). “If integrated diagnostics deliver greater value in cancer care, then adoption may be accelerated through value-based care models,” Malin observed. She said that bundled payments are one option for value-based payment, but she added that decisions then need to be made internally regarding how payments will be allocated.
Elenitoba-Johnson noted that lack of insurance coverage discourages the use of new technologies and that even when coverage exists, the reimbursement amount might not cover the costs. Malin highlighted the need to demonstrate the value of these technologies for patient care and outcomes and emphasized the importance of “being very clear about when are you providing value to the patient versus when is there some other value associated with [the technology].”
In response to a question about continued coverage of an evolving technology, Malin said that once a threshold of benefit has been realized and it has been established that the technology warrants coverage, it is generally not withdrawn, except in situations where there has been patient harm. Additional studies are needed when seeking coverage for additional use cases or populations, she said. Sawyers and Malin discussed how additional studies of diagnostic tests that have evolved over time might be paid for. Sawyers suggested that payers have a vested interest in generating evidence and might contribute
funding to support this research. Malin responded that payers contribute to the funding for the Patient-Centered Outcomes Research Institute.65
ENSURING PATIENT ACCESS AND PROMOTING HEALTH EQUITY
Many speakers discussed mechanisms to enable broad, equitable patient access to safe and effective integrated diagnostics, particularly in community-based settings of cancer care. The focus needs to be on “making sure that there is health equity in every one of our novel innovations, including integrated diagnostics” emphasized Beth Karlan, the Nancy Marks Endowed Chair in Women’s Health Research, and director of Cancer Population Genetics at the University of California, Los Angeles, Jonsson Comprehensive Cancer Center.
Developing Equitable AI Algorithms
Barzilay discussed challenges and solutions for developing safe and equitable AI algorithms. She pointed out that while there have long been disparities in health and biases in standard prediction models, there are several reasons to pay particular attention to the potential for bias in deep learning models. First, these models are “data hungry,” requiring a training set of 50,000–100,000 images, she said. For many clinical specialties, this volume of training data will not be available. Deep neural networks are also poor at handling distributional shifts in data. As an example, she showed how the performance of a model trained to recognize white numbers on black backgrounds declines when it is presented with colored numbers on different backgrounds. In addition, deep learning models can learn and perpetuate biases in the training dataset. She described how an image recognition model predicted that a monkey with a guitar was a human because the guitars it saw in the training data were always associated with humans.
“Most diagnostic tasks are beyond human prediction capacity,” Barzilay said. A human cannot look at an image and predict when someone will get cancer or have a recurrence, for example. She explained that this presents a problem for validating ML risk prediction models. She described three novel algorithmic solutions used to develop two validated risk prediction models, Mirai and Sybil, to ensure the models are robust. Mirai uses a patient’s screening mammogram to predict their risk of breast cancer within 5 years (Yala et al., 2021, 2022a, 2022b), and Sybil uses low-dose CT chest scans to predict a patient’s risk of developing lung cancer within 6 years (Mikhael et al., 2023).
___________________
65 See https://www.pcori.org/ (accessed January 28, 2024).
Anti-Bias Training Solution: Learning Invariant Representations
As discussed earlier, a model trained and validated in one population can perform very differently in another. Training with diverse data can help an algorithm like Mirai perform as intended across populations. However, if a training dataset contains a minority population, the algorithm will attempt to optimize for the majority dataset, leading to underperformance on the minority dataset, Barzilay said. The solution, she said, was “forcing the algorithm to see the majority and minority … in the same way, learning the representation directly, so it eliminates unnecessary differences between them and focuses on improving accuracy across this population.”
Distributional Shift Detection Solution: Learning to Split for Automatic Bias Detection
Barzilay said that another challenge is that the existence of bias in a training dataset is often unknown. This can result in the algorithm learning from the wrong information (Zech et al., 2018). Approaches to splitting datasets for training and testing vary and accuracy can vary depending on approach. Barzilay and colleagues developed an algorithm that learns to split the data for automatic bias detection, identifying outliers and out-of-distribution samples.66
Uncertainty Quantification Calibration Solution: Out of Domain Calibration
Barzilay expressed concern about popular misconceptions around transparent models and explainable AI and referred participants to a publication on the danger of believing “that we can use this interpretable model to improve the adoption in clinical systems” (Ghassemi et al., 2021). If a pathologist or radiologist is unable to make predictions about the location of future cancer recurrence, she asked, “how can [they] validate what the machine is doing?”
Barzilay explained that she and her colleagues developed new ML algorithms that use calibrated selective classification. In this approach, the model learns to abstain from making a prediction in the face of calibrated uncertainty.67 In essence, it is learning to say, “I don’t know,” she said. Instead of attempting to understand exactly how the algorithmic prediction is made, the algorithm reports when its prediction should not be trusted.
___________________
66 See Bao, Y., and R. Barzilay. 2022. Learning to Split for Automatic Bias Detection. https://arxiv.org/pdf/2204.13749.pdf (open access/pre-print repository document) (accessed May 26, 2023).
67 See Fisch et al. 2022. Calibrated Selective Classification. https://arxiv.org/pdf/2208.12084.pdf (open access/pre-print repository document) (accessed May 26, 2023).
“There are algorithmic solutions for safe and equitable AI deployment,” Barzilay concluded. More so than in other fields, ML models for clinical care need to be safe, work in different populations, and determine the certainty with which predictions can be trusted, she said.
Barzilay said the important indicator is how the algorithm performs compared to the current standard of care predictive models. “If you can better identify a population at risk, there is absolutely no reason not to use them,” she said. She noted that clinical trials in breast and lung cancer are underway to assess the extent to which an increase in predictive accuracy impacts long-term patient outcomes.
Integrated Diagnostics in Community-Based Settings of Care
Eighty-five percent of people in the United States receive their cancer care in community settings, said Randall Oyer, clinical professor of medicine at the Perelman School of Medicine and executive medical director of the Ann B. Barshinger Cancer Institute and Cancer Services at Penn Medicine Lancaster General Health. Oyer noted the uneven distribution of NCI-designated comprehensive cancer centers across the country,68 resulting in certain areas having more limited access to the technological advances available in these cancer centers.
Oyer noted that cancer care in the United States is generally provided in four settings: academic and research institutions (including comprehensive cancer centers), community practice, small private practices, and government settings (see Table 1). Oyer contrasted some of the main characteristics of academic research institutions and community practices, highlighting the differences while acknowledging the many overlaps. Academic research institutions “relentlessly pursue cure through research, teaching, and patient care,” he said, and the care workforce comprises heavily basic and clinical researchers focused on advancing research and providing highly specialized, cutting-edge care. The mission of community practice is population health and patient care, and the care workforce is generally oncology clinicians who provide the continuum of care in the community. Each has limitations, he said, such as the distance many patients must travel to reach an academic center and the varying depth of clinical specialization and availability of clinical trials in community practices.
Progress in oncology care is driven by innovation in technology, Oyer said. He outlined some of the technology milestones over the last 85 years that have contributed to advancing cancer care, including radiation therapy,
___________________
68 See https://www.cancer.gov/research/infrastructure/cancer-centers/find (accessed May 26, 2023).
TABLE 1 Settings of Cancer Care in the United States
| Academic/Research | Community Practice | Small/Private | Government | |
| Mission | Care research Teaching Patient care |
Population health Patient care |
Patient care Financial stability |
Total care for beneficiaries |
| Services (Majority) | Clinical trials Cancer care Technology |
Cancer care spectrum Clinical trials |
Medical oncology | Basic cancer care |
| Workforce | Basic & clinical researchers | Oncology clinicians | Medical oncologists | Oncology clinicians |
| Patients | Ability to travel Activated |
Entire spectrum | Privately insured | Government benefit/insurance |
| Highlights | Super specialized cutting-edge care | Continuum of care close to home | Familiar doctor-patient relationship | Defined benefits |
| Limitations | Distance Capacity Timelines |
Depth of specialization Availability of clinical trials |
Specialties beyond medical oncology Availability of clinical trials |
Timelines Availability of services |
SOURCE: Randall Oyer presentation, March 7, 2023.
chemotherapy, infusion pumps, genetics, telemedicine, and AI. He also acknowledged key organizations and structures that have helped to advance the field, including the NCI in 1937, National Cancer Act in 1971, medical oncology as a specialty (in the 1970s and 1980s), Community Clinical Oncology Program in 1983, Medicare Modernization Act, NCORP in 2011, and now integrated diagnostics.
Oyer emphasized the potential of integrated diagnostics to advance cancer care but said that “less than 15 percent of research is translated into practice,” and translation can take 17 years on average (Jørgensen, 2022). Some of the adoption challenges include cultural resistance to change, resource limitations, clinician education and training needs, and the operationalization of new prac-
tices within organizations. He suggested that that a new program or institute might be needed to drive progress by organizing the many laboratories and people around “a shared mission, a shared language, shared resources, [and] shared time lines.”
Oyer referred to a previous National Cancer Policy Forum workshop in which participants discussed the growing challenges facing the cancer careforce and potential solutions focused on improving patient experience and outcomes, the capacity and effectiveness of caregivers, and the efficiency, effectiveness, and resilience of clinicians (NASEM, 2019; Takvorian et al., 2022). Oyer said these solutions include “leveraging technology to share and integrate information to improve outcomes, efficiency, and reach,” and this includes digital technology and AI.
Patients receive care from different teams of clinicians, at different locations, using different tools, as they traverse the care continuum from diagnosis to treatment to surveillance and recovery, Oyer noted. He stressed the importance of precise, accurate, and timely bidirectional communication about integrated diagnostics across this continuum.
Oyer offered several suggestions to activate integrated diagnostics in community settings, such as:
- Establishing networks of care that share competence and collaboration across locations.
- Building infrastructure for cancer imaging networks to share expertise.
- Providing reports to clinicians that are timely, understandable, and usable in the clinic and at the bedside.
- Seamlessly incorporating integrated diagnostic reports into the EHR, and automatically interface them with widely available and widely used cancer care guidelines.
Oyer added that in order to ensure equity and representation, members of the cancer research community need to collaborate and build knowledge by collecting and sharing patient data, including the creation of a data bank, similar to the collection of tissue for a tissue bank.
Oyer also offered several suggestions to facilitate equitable access to precision diagnosis and clinical trials that improve outcomes for every patient with cancer in every community, such as expanding the capacity and representativeness of the oncology workforce, including diagnosticians. He added that digital tools can be used to augment capacity and that training is needed to ensure that caregivers can integrate these tools. Oyer also called on NCI and Congress to establish a “hub-and-spoke” system for the delivery of cancer care and to mandate EHR interoperability using mCODE.
Promoting Health Equity Through Patient Access to Integrated Diagnostics
“Whenever we introduce a new technology into medicine, we actually create disparities,” observed Otis Brawley, Bloomberg Distinguished Professor of Oncology and Epidemiology at the Johns Hopkins Bloomberg School of Public Health, and associate director of community outreach and engagement at the Johns Hopkins University Sydney Kimmel Comprehensive Cancer Center. Some people will have access to this technology, and others will not.
As background, Brawley said that U.S. spending on health care in 2019 was $3.8 trillion, which is 17.7 percent of the U.S. economy (Martin et al., 2021). For 2019, the United States ranked first in health care spending as a percentage of gross domestic product globally, followed by Germany at 11.7 percent. Despite high levels of health care spending, disparities in health persist and tend to be worse than those in other countries, he said, especially for mortality.
A disparity in health, Brawley said, is when a population fares worse than others in some measure (e.g., incidence, survival, quality of life, mortality). Characteristics used to define a population include sex, gender, race, ethnicity, culture, geographic origin, family or tribe, area of residence, and socioeconomic status. He noted the need for caution when defining a population by race. The U.S. Office of Management and Budget updates its definition of race every 10 years based on sociopolitical, not biological, characteristics. As such, race has changed over time.
Many Americans receive “less than optimal cancer care,” Brawley said, but this is more likely for racial minorities and people with low socioeconomic status. Subpar care can be a result of inaccessible, inadequate, or inappropriate screening, diagnostics, treatment, or therapy. These populations are also unlikely to have access to the newest tools and technologies in cancer care. Brawley pointed out, for example, that “Black Americans and poor Americans who need radiation therapy are more likely to be radiated by older, lower-energy, lower-quality radiation therapy machines compared to middle- and upper-middle-class Americans” (Mattes et al., 2021). In another example, he showed that disparities in colorectal cancer mortality between White and Black people emerged following implementation of screening and improvements in treatment in the 1970s (Siegel et al., 2018). Brawley said disparities in benefit from advances in screening and treatment are also seen in other cancers (e.g., breast cancer) and other diseases (e.g., cardiovascular disease and coronary artery bypass surgery). He also observed that “When we talk about health disparities, we have a substantial number of people who already get the diagnostics and then don’t get the treatment.”
There can also be disparities in the quality of pathological examination. For example, Brawley said that Black patients with colon cancer “are more likely to be treated in hospitals where the pathologist has multiple cases per day to process.” On average, minority patients with colon cancer have fewer lymph nodes assessed, which is associated with increased mortality (Rhoades et al., 2012). Inadequate examination of lymph nodes is associated with socioeconomic status (i.e., the hospital where a patient receives care). He said that because of this disparity in lymph node assessment, some Black patients are inaccurately diagnosed with stage 2 when they have stage 3 cancer, which has a higher risk of mortality. Because of this higher mortality rate, colon cancer has long been thought to be more aggressive in Black people than White people, but these errors in staging due to inadequate lymph node assessment could explain the disparities in outcomes.
The challenge, Brawley emphasized, is determining how to “provide adequate, high-quality care to populations who so often do not receive it.” Many health care settings in the United States are operating with limited resources, which are further consumed by unnecessary care. He said it is important to consider how new technologies can be implemented in resource-poor settings without further worsening disparities in care and outcomes. This is where implementation science can be applied to promote equitable uptake of new technologies, Brawley noted (Eccles and Mittman, 2006).
Drawing from his experience as director of a cancer center in a community safety net hospital, Brawley said that implementing lung cancer screening in a resource-poor setting, for example, can result in increased wait times for CT scans, thereby worsening overall quality of care for all patients. There can also be uneven implementation of new treatments and technologies. As examples, Brawley said the uptake of docetaxel69 for prostate cancer treatment was slower for older, poorer, and Black patients (Unger et al., 2014), and genetic testing for breast cancer gene mutations is underutilized among minority populations (Levy et al., 2011). A study of genetic testing for hereditary breast and ovarian cancers found that Asian and non-Hispanic Black patients were also more likely to have variants of unknown significance identified (Chapman-Davis et al., 2021). Brawley associated this inability to interpret genetic variants with the current lack of diversity in genome-wide association studies.
In addition to applying implementation science to advance the uptake and diffusion of new tools and technologies, Brawley said it is also important to deimplement (reduce or eliminate the use of) “inappropriate, ineffective, or potentially harmful health care services” (Walsh Bailey et al., 2021). For patients with breast cancer, for example, he said the advent of estrogen recep-
___________________
69 See https://www.cancer.gov/about-cancer/treatment/drugs/docetaxel (accessed September 5, 2023).
tor testing made it possible to stop treatment with tamoxifen70 for those who would not benefit from it. Brawley also emphasized the need for large clinical studies with subset analysis to elucidate the distribution of various markers among different types of populations (i.e., not only by race). Integrated diagnostics supports evidence-based care and prevention and “the rational practice of medicine,” he concluded.
Understanding and Addressing Disparities in Access and Use
Panelists were asked what could done to reduce disparities in access to diagnostics. Brawley and Oyer both said that the medical system cannot solve this problem alone. Many health disparities are rooted in “the fabric of society,” Brawley explained, and both agreed that political action is needed, but political will is lacking. Oyer said clinicians can work to bring new tools and technologies to their communities and “provide [care] to the patient in front of us, without bias, each and every day.” He advocated for a national universal health care system to help achieve this goal. Barzilay reiterated the need for diverse datasets that are representative of the whole population and stressed the importance of transparency when publishing studies based on biased historical datasets. She also said that the National Science Foundation (NSF) and NIH should dedicate sufficient resources to algorithm development to ensure that forthcoming AI systems perform well across different populations, especially minority populations.
Winn asked what infrastructure and workforce are needed to address the “data deserts” and how a “two-tiered AI system” might be avoided so that everyone receives the full benefit of the technology, not just those with access to an academic medical center. Brawley said that for many clinical settings, especially safety net hospitals, incorporating new technology and services can make it even harder to meet current needs of the community. He reiterated his example of how implementing lung cancer screening in safety net hospitals would benefit some people but would make the wait time for any type of CT scan longer for everyone, reducing quality of care. Brawley stressed that resources need to be provided for screening so that current services are not undermined.
Shulman pointed out that most smaller hospitals in rural areas are not part of a hub-and-spoke or other network, and these hospitals often have the largest disparities in care. Drawing on Winn’s comment about data deserts, Oyer and Shulman noted that said these rural areas are “deserts” outside of the catchment areas of NCI-designated cancer centers and called for a national effort to
___________________
70 See https://www.cancer.gov/publications/dictionaries/cancer-terms/def/tamoxifen-citrate (accessed September 5, 2023).
address these gaps. Shulman suggested that large academic medical centers have a responsibility to partner with these hospitals to improve care. Oyer suggested a role for NCI-designated cancer centers in expanding access to programs and digital resources at these hospitals. He called for bold and transformative solutions to ensure that these parts of the country have access to care.
Barzilay said there is a role for AI in taking a “more algorithmic approach of assessing risk and acting upon it.” Oyer said that health-related social needs should also be considered in patient risk assessment. They discussed the role of risk assessment in managing utilization of integrated diagnostics, ensuring access for patients who need a diagnostic test while reducing unnecessary testing of those who do not, especially in the face of limited resources for testing. Malin noted the importance of taking the intended use of the diagnostic test results into consideration. She agreed that not everyone needs to have every test done and added that unnecessary testing is costly for both the health system and the individual patient, emphasizing that the costs for patients include time spent on testing. Malin added that patient preferences need to be taken into consideration when deciding how comprehensive testing needs to be. For example, certain tests might not be necessary if a patient does not want to pursue a particular therapy.
CURRENT FEDERAL INITIATIVES IN PRECISION CANCER CARE
Two keynote speakers discussed current initiatives at NCI and the Advanced Research Project Agency for Health (ARPA-H) to advance precision oncology care.
NCI Precision Medicine Cancer Trials
Lyndsay Harris, associate director of the Cancer Diagnosis Program in the NCI Division of Cancer Treatment and Diagnosis, described the NCI Molecular Analysis for Therapy Choice (NCI-MATCH) clinical trial (Box 4).71 NCI-MATCH is a precision medicine trial that examines patient treatment approaches based on the molecular profiles of their tumors, said Harris. Data from the NCI-MATCH trial are being used to identify and evaluate potential diagnostic biomarkers.
NCI is also interested in studying how socioeconomic and environmental factors correlated with health disparities are associated with tumor and clinical
___________________
71 See https://www.cancer.gov/about-cancer/treatment/clinical-trials/nci-supported/nci-match (accessed May 26, 2023).
BOX 4
Overview of the NCI-MATCH Precision Cancer Medicine Trial
- Trial population: Adults with advanced solid tumors, lymphoma, or myeloma who are not responding to standard treatment or for whom there is no standard treatment.
-
Integrated workflow:
- Biopsy sample sequencing. Initially, standardized biopsy collection kits were sent to physician’s offices. Samples were sent to a central pathology laboratory for processing and then to one of four central laboratories for targeted panel sequencing. Later, to address low patient accrual for several arms, a designated laboratory network was set up to leverage next-generation sequencing of biopsy samples already being done as standard of care testing in many places. Laboratories then alerted physicians that their patients might be eligible for NCI-MATCH.
- AI-driven MATCHBox. Sequencing results were evaluated by MATCHBox, an AI-driven system that integrated information about the patient’s tumor gene abnormalities and other clinical information (e.g., histology) and determined whether they had a mutation that matched one of the 38 trial arms.
- Assignment to treatment. MATCHBox results were evaluated by a central review committee to confirm mutations of interest and treatment arm assignment for potential enrollment in the trial.
SOURCE: Lyndsay Harris presentation, March 7, 2023.
characteristics, Harris said. Data drawn from NCI-MATCH are being used to study, for example:
- The distribution of correlates of health disparities across the NCI-MATCH cohort;
- The association of a patient’s clinical status, diagnosis, and treatment history with correlates of health disparities;
- Whether potentially modifiable risk factors, such as excess body weight or smoking, differ by correlates of health disparities; and
- Whether there are any associations of correlates of health disparities with tumor molecular characteristics.
NCI is launching several large new precision medicine trials, including ComboMATCH,72 which is testing combinations of therapies selected on the basis of genomic testing results; MyeloMATCH,73 a multitiered study applying the NCI-MATCH strategy to acute myeloid leukemia and myelodysplastic syndromes; and iMATCH, studying targeted immunotherapy based on tumor molecular features. Harris said a new Precision Medicine Analysis and Coordination Center74 is responsible for an AI-driven treatment arm assignment and a range of other tasks (e.g., sequencing pipeline configuration; multiassay integration; integration with laboratory and clinical systems; biospecimen tracking; parsing, annotating, and molecular reporting; automated patient management workflows; treatment protocol management and tracking; data analytics, visualization, and reporting; support for protocol development and actionable alteration, identification, review, and curation; and support for the NCI-designated laboratory network).
ARPA-H and Cancer Moonshot
Susan Coller Monarez, deputy director of ARPA-H, provided a brief overview of the agency (see Box 5) and discussed its role in Cancer Moonshot. President Biden announced the creation of ARPA-H in 2022, stating that the agency “will pursue ideas that break the mold on how we normally support fundamental research and commercial products in this country.”75 The mission of ARPA-H is to “accelerate better health outcomes for everyone,” Monarez said, and the agency is focused on growing a portfolio of programs that will create transformational—rather than incremental—improvements in health through the development and commercialization of solutions.
Monarez underscored the pivotal role of ARPA-H program managers in driving the agency’s funded research portfolio.76 She said they aim to fund programs that use new approaches to solve a problem, with consideration of
___________________
72 See https://ecog-acrin.org/clinical-trials/eay191-combomatch/ (accessed February 2, 2024).
73 See https://www.cancer.gov/about-cancer/treatment/nci-supported/combomatch (accessed February 2, 2024),
74 See https://deainfo.nci.nih.gov/advisory/fac/1019/Doroshow.pdf (accessed February 2, 2024).
75 See https://www.whitehouse.gov/briefing-room/speeches-remarks/2022/03/18/remarks-by-president-biden-before-a-discussion-with-researchers-and-patients-on-advanced-research-project-agency-for-health-arpa-h/ (accessed May 26, 2023).
76 Monarez said that ARPA-H is actively seeking people to be program managers and fill other agency positions, and she encouraged workshop attendees to visit https://arpa-h.gov/careers/work-with-us/ for more information (accessed May 26, 2023).
BOX 5
Overview of ARPA-H
Features of ARPA-H
- Independent federal research and development funding agency reporting to the Secretary of the U.S. Department of Health and Human Services and situated within the National Institutes of Health.
- A $2.5 billion operating budget with a flexible funding structure to support high-risk, high-impact external research through cooperative agreements, other transactions authorities, and contracts (i.e., the agency is not grants based).
- Lean and nimble management structure that is capped at 210 federal employees.
- Research portfolio driven by program managers (appointed for a 3-year term with one renewal allowed) who identify high-impact, health-related problems to be addressed; solicit proposals and enter into agreements to fund the development of solutions; and define metrics and monitor progress toward milestones.
- Program managers developing proposals for program funding around a set of questions for evaluating proposed programs (see below)
ARPA-H Program Life Cycle
- Design. Identify challenging and well-defined problems and develop programs using the ARPA-(H)eilmeier questions and incorporating relevant input.
- Build. Broadly solicit proposals and build best-in-class performer teams to solve the problem.
- Perform. Program managers oversee program execution and performance against metrics, engaging experts throughout, and redirecting resources as needed.
- Learn. Continuously harness and apply knowledge across the program life cycle to enable transformational change.
- Transition. Build commercialization planning into a program at the start. Support licensing of solutions or company formation, mentor performers and create connections to investors and consumers, and derisk investment in solutions.
The ARPA-H Questions
- What are you trying to do? What health problem are you trying to solve?
- What are the current approaches to this problem? Who is involved? What are the limitations of present approaches?
- What is new about the approach to this problem? Why can it be successful at this time?
- Who cares? If successful, what difference will it make?
- What are the risks that may prevent achievement of the objectives? Any risks the program itself may present?
- How long will it take?
- How much will it cost?
- What are the mid-term and final exams to check for success?
- To ensure equitable access for all people, how will cost, accessibility, and user experience be addressed?
- How might this program be misperceived or misused (and how can that be prevented from happening)?
SOURCE: Susan Coller Monarez presentation, March 6, 2023.
why an effort to address this problem could be successful now and how the solution will ensure equitable access.
ARPA-H has four focus areas in which to build out its initial portfolio:
- Health Science Futures is focused on “accelerating advances across research areas and removing limitations that stymie progress,” Monarez said. Projects might involve broadly applicable tools and platforms, for example.
- Scalable Solutions will develop equitable solutions for overcoming challenges presented by geography, distribution, manufacturing, or data and information.
- Proactive Health will focus on prevention and wellness, including new approaches to disease detection and risk characterization and preventative interventions for a range of health threats.
- Resilient Systems is focused on creating the capabilities and business models needed to support integrated, resilient, sustainable health care systems that can also step up to meet the challenges of emerging crises, such as pandemics, social disruption, climate change, or economic instability.
Cancer Moonshot
ARPA-H is committed to supporting the Cancer Moonshot initiative, Monarez said. The agency is appointing a Cancer Moonshot Champion who will identify internal efforts across ARPA-H that are aligned with the initiative, engage members of the cancer research and care communities on behalf of ARPA-H, and collaborate with Cancer Moonshot leaders across government. Program managers who are pursuing solutions to cancer-related problems prioritized in the Moonshot can bring the resources of ARPA-H and its partners to bear (e.g., infrastructure, implementation pathways) and “translate ongoing research efforts into capabilities for researchers or patients,” she said.
Monarez said some of the areas in which ARPA-H programs could address Moonshot strategic priorities include “closing the screening gaps, addressing environmental exposures, decreasing the impact of preventable cancers, supporting patients and caregivers, bringing cutting-edge research advances to patients, and addressing inequities.” She cited advancing digital histopathology capabilities as a specific example of a potential ARPA-H program that would be aligned with the Moonshot strategic priority to bring cutting-edge research advances to patients. ARPA-H supported research might also explore the development of novel multiomic histopathology assays; the use of AI and ML to expand automation in histopathology practice; and integrating histopathology data into clinical care pathways.
Arunan Skandarajah, presidential innovation fellow at ARPA-H, encouraged workshop participants to propose projects to ARPA-H and said that agency staff also welcome conversations with individuals or teams about potential integrated diagnostics concepts that might be suitable for an ARPA-H program.
Promoting a Culture of Data Sharing
Several speakers asked about the extent to which the NCI and ARPA-H initiatives would be making data publicly available. Harris noted that NIH recently updated its data sharing policy77 and under the new policy, “all molecular data and clinical data must be shared at the end of the study.” Data from NCI grantees and other NCI programs are accessible through the Cancer Research Data Commons.78 Monarez said ARPA-H is developing its data sharing policies, which will be in alignment with the Public Access Policy
___________________
77 See https://sharing.nih.gov/data-management-and-sharing-policy/about-data-management-and-sharing-policies/data-management-and-sharing-policy-overview#after (accessed January 31, 2024).
78 See https://datascience.cancer.gov/data-commons (accessed September 5, 2023).
of the White House Office of Science and Technology Policy.79 The intent is that all data will be “available to those who would benefit from its utilization.” “Without data, you can’t innovate,” she concluded.
REFLECTING ON THE WORKSHOP DISCUSSIONS
In the final session, the moderators of each session reflected on key points highlighted by speakers, including opportunities to advance the use of integrated diagnostics in cancer care (see also Boxes 1 and 2).
Envisioning the Future for Integrated Diagnostics
Hricak emphasized that “diagnostics are moving toward data science, and there is a need for data integration.” She suggested that the scope of integrated diagnostics remain flexible and patient centered, noting that these go beyond a clinical decision support system or data display dashboard. Many speakers highlighted that funding for integrated diagnostics can be a challenge, and it can be difficult to demonstrate a return on investment in the short term, Hricak said. Elenitoba-Johnson called for the development of policies and incentives to promote the adoption of integrated diagnostics in clinical practice. Hricak said other considerations highlighted by many speakers included improving interoperability of data systems across clinical disciplines; implementing data standards, synoptic operative reports, and a lexicon to convey the degree of diagnostic certainty; facilitating collaboration to transition image annotation and segmentation from manual to automated; fostering a culture of data sharing; restructuring reimbursement to cover the diagnosis versus diagnostic testing procedures; scaling up precision diagnostics to reach broader populations; and leveraging institutional governance to adopt integrated diagnostics within clinical care.
Highlighting Insights from Academia, Industry, and Health Care Organizations
Nancy Davidson, executive vice president for Clinical Affairs at the Fred Hutchinson Cancer Center and Raisbeck Endowed Chair for Collaborative Cancer Research at the University of Washington, highlighted perspectives from representatives of academic medical centers, industry, and large health care organizations. Several speakers stressed the need to facilitate the integra-
___________________
79 See https://www.whitehouse.gov/wp-content/uploads/2022/08/08-2022-OSTP-Public-Access-Memo.pdf (accessed February 1, 2024).
tion of data from across disciplinary or organizational silos, she said, while others suggested that integrated diagnostics could better manage and communicate diagnostic testing results. To promote adoption, several speakers suggested conducting analyses to assess the value of integrated diagnostics in achieving high-quality, efficient care, which could provide evidence to support payment models and insurance coverage.
Davidson said that many speakers highlighted the importance of training the next generation of clinicians to take an integrated approach to diagnostics and to train specialists in integrated diagnostics. Davidson added that much could be learned from health systems that are implementing and scaling integrating diagnostics. Some of the lessons shared by speakers included facilitating structured reporting to integrate data across diagnostic specialties, ensuring feedback loops to assess diagnostic performance, and ensuring that a primary focus of integrated diagnostics is patient-centered care.
Improving Evidence Generation
Much of the discussion on improving evidence generation for integrated diagnostics centered on the opportunities to use EHR data and capabilities to support the evaluation of integrated diagnostics, Levy said, adding that this includes working with EHR vendors to advance and disseminate use cases. Many speakers said that EHR data collected during routine care could be leveraged for pragmatic studies and suggested being more intentional about collecting EHR data for prospective clinical trials. Levy said that speakers highlighted the need for “significant investment in evidence generation for analytic validity, clinical validity, and clinical utility” to support regulatory review and implementation of integrated diagnostics in clinical practice. Although challenges persist, “the culture of data sharing for discovery remains strong,” Levy stressed. She concluded that “regulations, policies, and guidelines that recommend minimum structured documentation standards for diagnostic reporting influence documentation practices that enables evidence generation for integrated diagnostics.”
Enhancing Design and Use of Integrated Diagnostics
Jensen said one of the recurring topics of discussion around design and use of integrated diagnostics was the potential for AI algorithms to perpetuate and amplify biases in training datasets, with many speakers highlighting the importance of training and testing models on datasets that reflect the diversity of the population. This includes ensuring that patient enrollment in clinical trials of integrated diagnostics is diverse and inclusive, which also expands patient access to cutting-edge research, because these prospective trial
populations are likely to be used for future research, including AI algorithm development.
Jensen said another topic was the use of DMTs to provide guidance to treating clinicians on test ordering and interpreting test results. Jensen said it might be helpful to develop a DMT on specific clinical areas at the outset and expand capacity to additional areas, rather than attempting a comprehensive rollout. Jensen also suggested that research funders support development of DMTs.
Naik and Jason Slagle, research associate professor in the department of anesthesiology at Vanderbilt University Medical Center, highlighted additional design and use considerations that speakers discussed, including the need for investments in data engineering to scale the integration of data from across the diagnostic specialties. Slagle said that successful implementation of integrated diagnostics also hinges on trust, understanding, accountability, timing options, prioritization, annotation, and actionability. He added that these issues affect clinicians’ perceptions of integrated diagnostics and their willingness to use them. Naik pointed out that once implemented, ongoing testing should incorporate “the principles of high reliability organizations to ensure reliability and reduce unwanted variation.” He also noted the need for open-source protocols for algorithms and interinstitutional sharing and testing of protocols to support scalability and sustainability.
Many speakers emphasized the need to redesign EHRs to better meet clinician needs and facilitate integrated diagnostics, Slagle said. Some of the suggested changes included incorporating data visualization, multilevel modeling, and ML approaches for workflow and diagnostics, he summarized. Using principles of risk communication to support shared decision making was also discussed as an opportunity to improve patient care. Naik added that some speakers suggested a code of conduct for implementing AI-based integrated diagnostics and also leveraging innovative clinical trial designs to facilitate simultaneous testing of tools and implementation strategies. Finally, Slagle noted that more data scientists need to be recruited to the fields of cancer research and care.
Regulatory Oversight and Insurance Coverage
Jensen suggested that developers of integrated diagnostics meet with FDA leadership to discuss regulatory pathways to evaluate them. In regard to insurance coverage, he noted “there seems to be a reticence … in paying for integrated diagnostics as a stand-alone component of medical care” and suggested that moving toward bundled payments for oncology care might partially address this issue.
Access and Equity
“Equity needs to be a part of all we do,” stressed Wendy Nilsen, acting deputy division director of the NSF Industrial Innovation and Partnerships Division. She noted that many speakers called attention to the complexity of AI-based algorithms and the need to validate them across institutions and populations. Several solutions used in the development of validated risk prediction models were discussed, including an approach to splitting datasets for training and testing in which the algorithm learns to detect bias automatically. The challenges of developing explainable AI were also discussed and a ML model that uses calibrated selective classification was described. As an alternative to trying to explain how an algorithm makes a prediction, this model learns to report when its prediction cannot be trusted based on calibrated uncertainty, she said.
The distribution of cancer care across the United States was discussed, noting clear disparities in access to high-quality care and new technologies, Nilsen said. Because most patients with cancer receive their care in the community, participants discussed the need for a hub-and-spoke system to better enable the equitable deployment of integrated diagnostics and the delivery of cancer care, and to build research on dissemination and implementation into product development, she summarized.
Nilsen noted that workforce issues were raised, along with the importance of multidisciplinary teams in developing, implementing, and using integrated diagnostics. Several participants discussed the need to leverage new digital technologies to “increase caregiver capacity, patient experience, and clinical efficiency and effectiveness” and the capacity and skills of the oncology workforce to integrate diagnostic tools and improve evidence generation.
Elenitoba-Johnson observed the lack of uniformity in how policies and payment models are deployed in different regions or care venues and how an organization’s market share impacts its negotiating power with payers. He said crosscutting policies will be needed to support the delivery integrated diagnostics at scale. Lennerz agreed but added that “local coverage policy can drive national coverage policy and vice versa.” Levy noted that a bundled payment model already exists for inpatient care, and that could be used to test the incorporation of integrated diagnostics.
Participants shared additional reflections on opportunities to advance technology, access, and equity. Shulman highlighted the need to bring in human-centered design from other industries (e.g., the airplane cockpit). The potential of integrated diagnostics to improve care and outcomes will not be realized if clinicians cannot readily use the resulting information during a patient visit, he said. Hricak agreed and noted that new radiology dashboard designs are much improved to display key clinical findings. Naik added that
user-centered design should address the needs of both the clinician and the patient as users. Karlan also emphasized the need to engage those users when developing new AI-based integrated diagnostics. Winn highlighted the difference between useful and usable. Something that is useful is “fit for a purpose” but not necessarily “fit to be used.” In the current context, data can be useful but not necessarily usable by different end users. The usefulness, and usability, of discoveries should be considered in parallel, he said, emphasizing that a critical disconnect in health is that there is a “lot of discovering, but that doesn’t cross the bridge to implementation,” and as a result, not everyone benefits from the advances. “That’s why I really think about the useful discovery, usefulness, and at the end, the usability,” he said.
REFERENCES
Acosta, J. N., G. J. Falcone, P. Rajpurkar, and E. J. Topol. 2022. Multimodal biomedical AI. Nature Medicine 28(9):1773–1784.
Asan, O., A. E. Bayrak, and A. Choudhury. 2020. Artificial intelligence and human trust in healthcare: Focus on clinicians. Journal of Medical Internet Research 22(6):e15154.
Boehm, K. M., E. A. Aherne, L. Ellenson, I. Nikolovski, M. Alghamdi, I. Vázquez-García, D. Zamarin, K. Long Roche, Y. Liu, D. Patel, A. Aukerman, A. Pasha, D. Rose, P. Selenica, P. I. Causa Andrieu, C. Fong, M. Capanu, J. S. Reis-Filho, R. Vanguri, H. Veeraraghavan, N. Gangai, R. Sosa, S. Leung, A. McPherson, J. Gao, MSK MIND Consortium, Y. Lakhman, and S. P. Shah. 2022a. Multimodal data integration using machine learning improves risk stratification of high-grade serous ovarian cancer. Nature Cancer 3(6):723–733.
Boehm, K. M., P. Khosravi, R. Vanguri, J. Gao, and S. P. Shah. 2022b. Harnessing multimodal data integration to advance precision oncology. Nature Reviews Cancer 22(2):114–126.
Bourla, A. B., and N. J. Meropol. 2021. Bridging the divide between clinical research and clinical care in oncology: An integrated real-world evidence generation platform. Digital Health 7:20552076211059975.
Burq, M., E. Rainaldi, K. C. Ho, C. Chen, B. R. Bloem, L. J. W. Evers, R. C. Helmich, L. Myers, W. J. Marks, Jr., and R. Kapur. 2022. Virtual exam for Parkinson’s disease enables frequent and reliable remote measurements of motor function. NPJ Digital Medicine 23;5(1):65. Erratum in NPJ Digital Medicine 5(1):196.
Carlsson, A., V. S. Nair, M. S. Luttgen, K. V. Keu, G. Horng, M. Vasanawala, A. Kolatkar, M. Jamali, A. H. Iagaru, W. Kuschner, B. W. Loo, Jr., J. B. Shrager, K. Bethel, C. K. Hoh, L. Bazhenova, J. Nieva, P. Kuhn, and S. S. Gambhir. 2014. Circulating tumor microemboli diagnostics for patients with non-small-cell lung cancer. Journal of Thoracic Oncology 9(8):1111–1119.
Chakravarty, D., J. Gao, S. M. Phillips, R. Kundra, H. Zhang, J. Wang, J. E. Rudolph, R. Yaeger, T. Soumerai, M. H. Nissan, M. T. Chang, S. Chandarlapaty, T. A. Traina, P. K. Paik, A. L. Ho, F. M. Hantash, A. Grupe, S. S. Baxi, M. K. Callahan, A. Snyder, P. Chi, D. Danila, M. Gounder, J. J. Harding, M. D. Hellmann, G. Iyer, Y. Janjigian, T. Kaley, D. A. Levine, M. Lowery, A. Omuro, M. A. Postow, D. Rathkopf, A. N. Shoushtari, N. Shukla, M. Voss, E. Paraiso, A. Zehir, M. F. Berger, B. S. Taylor, L. B. Saltz, G. J. Riely, M. Ladanyi, D. M. Hyman, J. Baselga, P. Sabbatini, D. B. Solit, and N. Schultz. 2017. OncoKB: A precision oncology knowledge base. JCO Precision Oncology 2017:PO.17.00011.
Chakravorti, B. 2022. Why AI failed to live up to its potential during the pandemic. Harvard Business Review. https://hbr.org/2022/03/why-ai-failed-to-live-up-to-its-potential-during-the-pandemic (accessed May 26, 2023).
Chapman-Davis, E., Z. N. Zhou, J. C. Fields, M. K. Frey, B. Jordan, K. J. Sapra, S. Chatterjee-Paer, A. D. Carlson, and K. M. Holcomb. 2021. Racial and ethnic disparities in genetic testing at a hereditary breast and ovarian cancer center. Journal of General Internal Medicine 36(1):35–42.
Choudhury, A. 2022. Toward an ecologically valid conceptual framework for the use of artificial intelligence in clinical settings: Need for systems thinking, accountability, decision-making, trust, and patient safety considerations in safeguarding the technology and clinicians. JMIR Human Factors 9(2):e35421.
Choudhury A., and O. Asan. 2022. Impact of accountability, training, and human factors on the use of artificial intelligence in healthcare: Exploring the perceptions of healthcare practitioners in the U.S. Human Factors in Healthcare 2:100021
Choudhury, A., and S. Elkefi. 2022. Acceptance, initial trust formation, and human biases in artificial intelligence: Focus on clinicians. Frontiers in Digital Health 4:966174.
Choudhury, A., O. Asan, and J. E. Medow. 2022. Clinicians’ perceptions of an artificial intelligence-based blood utilization calculator: Qualitative exploratory study. JMIR Human Factors 9(4):e38411.
Cui, C., H. Yang, Y. Wang, S. Zhao, Z. Asad, L. A. Coburn, K. T. Wilson, B. A. Landman, and Y. Huo. 2023. Deep multimodal fusion of image and non-image data in disease diagnosis and prognosis: A review. Progress in Biomedical Engineering 5(2).
Culver, J. O., J. L. Hull, D. F. Dunne, and W. Burke. 2001. Oncologists’ opinions on genetic testing for breast and ovarian cancer. Genetics in Medicine 3(2):120–125.
Dagogo-Jack, I., A. Manoogian, N. Jessop, N. Z. Georgantas, F. J. Fintelmann, A. Farahani, S. R. Digumarthy, M. C. Price, E. E. Folch, C. M. Keyes, A. Do, J. L. Peterson, M. Mino-Kenudson, M. Pitman, M. Rivera, V. Nardi, D. Dias-Santagata, L. P. Le, A. J. Iafrate, R. S. Heist, L. R. Ritterhouse, and J. K. Lennerz. 2023. Integrated radiology, pathology, and pharmacy program to accelerate access to osimertinib. JCO Oncology Practice 19(9):786–792.
Damschroder, L. J., C. M. Reardon, M. A. O. Widerquist, and J. Lowery. 2022. The updated Consolidated Framework for Implementation Research based on user feedback. Implementation Science 17(1):75.
de Haan, R. R., M. J. Schreuder, E. Pons, and J. J. Visser. 2019. Adrenal incidentaloma and adherence to international guidelines for workup based on a retrospective review of the type of language used in the radiology report. Journal of the American College of Radiology 16(1):50–55.
Dorr, D. A., R. L. Ross, D. Cohen, D. Kansagara, K. Ramsey, B. Sachdeva, and J. P. Weiner. 2021. Primary care practices’ ability to predict future risk of expenditures and hospitalization using risk stratification and segmentation. BMC Medical Informatics and Decision Making 21(1):104.
Eccles, M. P., and B. S. Mittman. 2006. Welcome to Implementation Science. Implementation Science 1:1.
Echle, A., H. I. Grabsch, P. Quirke, P. A. van den Brandt, N. P. West, G. G. A. Hutchins, L. R. Heij, X. Tan, S. D. Richman, J. Krause, E. Alwers, J. Jenniskens, K. Offermans, R. Gray, H. Brenner, J. Chang-Claude, C. Trautwein, A. T. Pearson, P. Boor, T. Luedde, N. T. Gaisa, M. Hoffmeister, and J. N. Kather. 2020. Clinical-grade detection of microsatellite instability in colorectal tumors by deep learning. Gastroenterology 159(4):1406–1416.
Egger, J., C. Gsaxner, A. Pepe, K. L. Pomykala, F. Jonske, M. Kurz, J. Li, and J. Kleesiek. 2022. Medical deep learning—a systematic meta-review. Computer Methods and Programs in Biomedicine 221:106874.
Elwyn, G., M. A. Durand, J. Song, J. Aarts, P. J. Barr, Z. Berger, N. Cochran, D. Frosch, D. Galasiński, P. Gulbrandsen, P. K. J. Han, M. Härter, P. Kinnersley, A. Lloyd, M. Mishra, L. Perestelo-Perez, I. Scholl, K. Tomori, L. Trevena, H. O. Witteman, and T. Van der Weijden. 2017. A three-talk model for shared decision making: Multistage consultation process. BMJ 359:j4891.
Emamekhoo, H., C. B. Carroll, C. Stietz, J. B. Pier, M. D. Lavitschke, D. Mulkerin, M. E. Sesto, and A. J. Tevaarwerk. 2022. Supporting structured data capture for patients with cancer: An initiative of the University of Wisconsin Carbone Cancer Center Survivorship Program to Improve Capture of Malignant Diagnosis and Cancer Staging Data. JCO Clinical Cancer Informatics 6:e2200020.
Foraker, R., C. Phommasathit, K. Clevenger, C. Lee, J. Boateng, N. Shareef, and M. C. Politi. 2023. Using the sociotechnical model to conduct a focused usability assessment of a breast reconstruction decision tool. BMC Medical Informatics Decision Making 23(1):140.
Gambhir, S. 2018. The role of the radiologist in 21st century healthcare. Presentation at the Joint Symposium on the Future of Medical Imaging: Trends and Perspectives. https://is3r.org/members-area/interim-meetings/2018-2/ (accessed May 26, 2023).
Ghassemi, M., L. Oakden-Rayner, and A. L. Beam. 2021. The false hope of current approaches to explainable artificial intelligence in health care. Lancet Digital Health 3(11):e745-e750.
Ghesu, F. C., B. Georgescu, A. Mansoor, Y. Yoo, D. Neumann, P. Patel, R. S. Vishwanath, J. M. Balter, Y. Cao, S. Grbic, and D. Comaniciu. 2022. Contrastive self-supervised learning from 100 million medical images with optional supervision. Journal of Medical Imaging 9(6):064503.
Gruber, T. 2016. Ontology. In Encyclopedia of database systems, edited by L. Liu and M. T. Özsu. New York, NY: Springer New York. Pp. 1–3.
Habli, I., T. Lawton, and Z. Porter. 2020. Artificial intelligence in health care: Accountability and safety. Bulletin of the World Health Organization 98(4):251–256.
Han, P. K. J., T. D. Strout, C. Gutheil, C. Germann, B. King, E. Ofstad, P. Gulbrandsen, and R. Trowbridge. 2021. How physicians manage medical uncertainty: A qualitative study and conceptual taxonomy. Medical Decision Making 41(3):275–291.
Hasak, J. M., T. M. Myckatyn, V. F. Grabinski, S. E. Philpott, R. P. Parikh, and M. C. Politi. 2017. Stakeholders’ perspectives on postmastectomy breast reconstruction: Recognizing ways to improve shared decision making. Plastic and Reconstructive Surgery Global Open 5(11):e1569.
He, X., X. Liu, F. Zuo, H. Shi, and J. Jing. 2023. Artificial intelligence-based multi-omics analysis fuels cancer precision medicine. Seminars in Cancer Biology 88:187–200.
Johnson, B. E., A. L. Creason, J. M. Stommel, J. M. Keck, S. Parmar, C. B. Betts, A. Blucher, C. Boniface, E. Bucher, E. Burlingame, T. Camp, K. Chin, J. Eng, J. Estabrook, H. S. Feiler, M. B. Heskett, Z. Hu, A. Kolodzie, B. L. Kong, M. Labrie, J. Lee, P. Leyshock, S. Mitri, J. Patterson, J. L. Riesterer, S. Sivagnanam, J. Somers, D. Sudar, G. Thibault, B. R. Weeder, C. Zheng, X. Nan, R. F. Thompson, L. M. Heiser, P. T. Spellman, G. Thomas, E. Demir, Y. H. Chang, L. M. Coussens, A. R. Guimaraes, C. Corless, J. Goecks, R. Bergan, Z. Mitri, G. B. Mills, and J. W. Gray. 2022. An omic and multidimensional spatial atlas from serial biopsies of an evolving metastatic breast cancer. Cell Reports Medicine 3(2):100525.
Jørgensen, J. T. 2020. Companion and complementary diagnostics: An important treatment decision tool in precision medicine. Expert Review of Molecular Diagnostics 20(6):557–559.
Lavery, J. A., E. M. Lepisto, S. Brown, H. Rizvi, C. McCarthy, M. LeNoue-Newton, C. Yu, J. Lee, X. Guo, T. Yu, J. Rudolph, S. Sweeney, AACR Project GENIE Consortium, B. H. Park, J. L. Warner, P. L. Bedard, G. Riely, D. Schrag, and K. S. Panageas. 2022. A scalable quality assurance process for curating oncology electronic health records: The Project GENIE Biopharma Collaborative Approach. JCO Clinical Cancer Informatics 6:e2100105.
Lee, C. N., J. Sullivan, R. Foraker, T. M. Myckatyn, M. A. Olsen, C. Phommasathit, J. Boateng, K. L. Parrish, M. Rizer, T. Huerta, and M. C. Politi. 2022. Integrating a patient decision aid into the electronic health record: A case report on the implementation of BREASTChoice at 2 sites. MDM Policy & Practice 7(2):23814683221131317.
Lennerz, J. K., U. Green, D. F. K. Williamson, and F. Mahmood. 2022. A unifying force for the realization of medical AI. NPJ Digital Medicine 5(1):172.
Lennerz, J. K., R. Salgado, G. E. Kim, S. J. Sirintrapun, J. C. Thierauf, A. Singh, I. Indave, A. Bard, S. E. Weissinger, Y. K. Heher, M. E. de Baca, I. A. Cree, S. Bennett, A. Carobene, T. Ozben, and L. L. Ritterhouse. 2023. Diagnostic quality model (DQM): An integrated framework for the assessment of diagnostic quality when using AI/ML. Clinical Chemistry and Laboratory Medicine 61(4):544–557.
Levy, D. E., S. D. Byfield, C. B. Comstock, J. E. Garber, S. Syngal, W. H. Crown, and A. E. Shields. 2011. Underutilization of BRCA1/2 testing to guide breast cancer treatment: Black and Hispanic women particularly at risk. Genetics in Medicine 13(4):349–355.
Lipkova, J., R. J. Chen, B. Chen, M. Y. Lu, M. Barbieri, D. Shao, A. J. Vaidya, C. Chen, L. Zhuang, D. F. K. Williamson, M. Shaban, T. Y. Chen, F. and Mahmood. 2022. Artificial intelligence for multimodal data integration in oncology. Cancer Cell 40(10):1095–1110.
Lippi, G., and M. Plebani. 2020. Integrated diagnostics: The future of laboratory medicine? Biochemia Medica 30(1):010501.
Liu, F., O. Vermesh, V. Mani, T. J. Ge, S. J. Madsen, A. Sabour, E. C. Hsu, G. Gowrishankar, M. Kanada, J. V. Jokerst, R. G. Sierra, E. Chang, K. Lau, K. Sridhar, A. Bermudez, S. J. Pitteri, T. Stoyanova, R. Sinclair, V. S. Nair, S. S. Gambhir, and U. Demirci. 2017. The exosome total isolation chip. ACS Nano 11(11):10712–10723.
Lou, B., S. Doken, T. Zhuang, D. Wingerter, M. Gidwani, N. Mistry, L. Ladic, A. Kamen, and M. E. Abazeed. 2019. An image-based deep learning framework for individualizing radiotherapy dose. Lancet Digital Health 1(3):e136–e147.
Marquart, J., E. Y. Chen, and V. Prasad. 2018. Estimation of the percentage of U.S. patients with cancer who benefit from genome-driven oncology. JAMA Oncology 4(8):1093–1098.
Martin, A. B., M. Hartman, D. Lassman, A. Catlin, and the National Health Expenditure Accounts Team. 2021. National health care spending in 2019: Steady growth for the fourth consecutive year. Health Affairs 40(1):14–24.
Martin, S., J. Wagner, N. Lupulescu-Mann, K. Ramsey, A. Cohen, P. Graven, N, G. Weiskopf, and D. A. Dorr. 2017. Comparison of EHR-based diagnosis documentation locations to a gold standard for risk stratification in patients with multiple chronic conditions. Applied Clinical Informatics 8(3):794–809.
Mattes, M. D., G. Suneja, B. G. Haffty, C. Takita, M. S. Katz, N. Ohri, C. Deville, Jr., M. L. Siker, and H. S. Park. 2021. Overcoming barriers to radiation oncology access in low-resource settings in the United States. Advances in Radiation Oncology 6(6):100802.
Messiou, C., R. Lee, and M. Salto-Tellez. 2023. Multimodal analysis and the oncology patient: Creating a hospital system for integrated diagnostics and discovery. Computational and Structural Biotechnology Journal 21:4536-4539.
Mikhael, P. G., J. Wohlwend, A. Yala, L. Karstens, J. Xiang, A. K. Takigami, P. P. Bourgouin, P. Chan, S. Mrah, W. Amayri, Y. H. Juan, C. T. Yang, Y. L. Wan, G. Lin, L. V. Sequist, F. J. Fintelmann, and R. Barzilay. 2023. Sybil: A validated deep learning model to predict future lung cancer risk from a single low-dose chest computed tomography. Journal of Clinical Oncology 41(12):2191–2200.
Nahum-Shani, I., S. D. Shaw, S. M. Carpenter, S. A. Murphy, and C. Yoon. 2022. Engagement in digital interventions. American Psychologist 77(7):836–852.
Nair, V. S., K. V. Keu, M. S. Luttgen, A. Kolatkar, M. Vasanawala, W. Kuschner, K. Bethel, A. H. Iagaru, C. Hoh, J. B. Shrager, B. W. Loo, Jr., L. Bazhenova, J. Nieva, S. S. Gambhir, and P. Kuhn. 2013. An observational study of circulating tumor cells and (18)F-FDG PET uptake in patients with treatment-naive non-small cell lung cancer. PLoS One 8(7):e67733.
NAM (National Academy of Medicine). 2017. Optimizing strategies for clinical decision support: Summary of a meeting series. Washington, DC: The National Academies Press.
NASEM (National Academies of Sciences, Engineering, and Medicine). 2015. Improving diagnosis in health care. Washington, DC: The National Academies Press.
NASEM. 2019. Developing and sustaining an effective and resilient oncology careforce. Washington, DC: The National Academies Press.
NASEM. 2022. Artificial intelligence in health care: The hope, the hype, the promise, the peril. Washington, DC: The National Academies Press.
Nguyen, T., P. Lai, N. H. Phan, and M. T. Thai. 2022. XRand: Differentially private defense against explanation-guided attacks. Presented at the 2023 AAAI Conference on Artificial Intelligence. https://arxiv.org/pdf/2212.04454.pdf (accessed May 26, 2023).
Obermeyer, Z., B. Powers, C. Vogeli, and S. Mullainathan. 2019. Dissecting racial bias in an algorithm used to manage the health of populations. Science 366(6464):447–453.
Ogier du Terrail, J., A. Leopold, C. Joly, C. Béguier, M. Andreux, C. Maussion, B. Schmauch, E. W. Tramel, E. Bendjebbar, M. Zaslavskiy, G. Wainrib, M. Milder, J. Gervasoni, J. Guerin, T. Durand, A. Livartowski, K. Moutet, C. Gautier, I. Djafar, A. L. Moisson, C. Marini, M. Galtier, F. Balazard, R. Dubois, J. Moreira, A. Simon, D. Drubay, M. Lacroix-Triki, C. Franchet, G. Bataillon, and P. E. Heudel. 2023. Federated learning for predicting histological response to neoadjuvant chemotherapy in triple-negative breast cancer. Nature Medicine 29(1):135–146.
Osterman, T. J., J. C. Yao, and M. K. Krzyzanowska. 2023. Implementing innovation: Informatics-based technologies to improve care delivery and clinical research. American Society of Clinical Oncology Educational Book (43):e389880.
Parikh, R., A. Mathai, S. Parikh, G. Chandra Sekhar, and R. Thomas. 2008. Understanding and using sensitivity, specificity and predictive values. Indian Journal of Ophthalmology 56(1):45–50.
Phan, N. H., M. N. Vu, Y. Liu, R. Jin, D. Dou, X. Wu, and M. T. Thai. 2019. Heterogeneous Gaussian mechanism: Preserving differential privacy in deep learning with provable robustness. In S. Kraus (Ed.), Proceedings of the 28th International Joint Conference on ArtificialIntelligence, 2019. Pp. 4753–4759. https://www.ijcai.org/proceedings/2019/0660.pdf (accessed May 26, 2023).
Phan, H., M. T. Thai, H. Hu, J. Ruoming, S. Tong, and D. Dejing. 2020. Scalable differential privacy with certified robustness in adversarial learning. Presented at the 37th International Conference on Machine Learning. https://arxiv.org/pdf/1903.09822.pdf (accessed May 26, 2023).
Politi, M. C., P. Adsul, M. D. Kuzemchak, R. Zeuner, and D. L. Frosch. 2015. Clinicians’ perceptions of digital vs. paper-based decision support interventions. Journal of Evaluation in Clinical Practice 21(2):175–179.
Politi, M. C., C. N. Lee, S. E. Philpott-Streiff, R. E. Foraker, M. A. Olsen, C. Merrill, Y. Tao, and T. M. Myckatyn. 2020. A randomized controlled trial evaluating the BREASTChoice tool for personalized decision support about breast reconstruction after mastectomy. Annals of Surgery 271(2):230–237.
Porter, M. E. 2010. What is value in health care? New England Journal of Medicine 363(26):2477–2481.
Pugh, T. J., J. L. Bell, J. P. Bruce, G. J. Doherty, M. Galvin, M. F. Green, H. Hunter-Zinck, P. Kumari, M. L. Lenoue-Newton, M. M. Li, J. Lindsay, T. Mazor, A. Ovalle, S. J. Sammut, N. Schultz, T. V. Yu, S. M. Sweeney, B. Bernard, and the AACR Project GENIE Consortium, Genomics, and Analysis Working Group. 2022. AACR Project GENIE: 100,000 cases and beyond. Cancer Discovery 12(9):2044–2057.
Randall, J., P. T. Teo, B. Lou, J. Shah, J. Patel, A. Kamen, and M. E. Abazeed. 2023. Image-based deep neural network for individualizing radiotherapy dose is transportable across health systems. JCO Clinical Cancer Informatics 7:e2200100.
Rani, V., S. T. Nabi, M. Kumar, A. Mittal, and K. Kumar. 2023. Self-supervised learning: A succinct review. Archives of Computational Methods Engineering: State of the Art Reviews 30(4):2761–2775.
Rasche, L., E. Angtuaco, J. E. McDonald, A. Buros, C. Stein, C. Pawlyn, S. Thanendrarajan, C. Schinke, R. Samant, S. Yaccoby, B. A. Walker, J. Epstein, M. Zangari, F. van Rhee, T. Meissner, H. Goldschmidt, K. Hemminki, R. Houlston, B. Barlogie, F. E. Davies, G. J. Morgan, and N. Weinhold. 2017. Low expression of hexokinase-2 is associated with false-negative FDG-positron emission tomography in multiple myeloma. Blood 130(1):30–34.
Rhoads, K. F., J. Cullen, J. V. Ngo, and S. M. Wren. 2012. Racial and ethnic differences in lymph node examination after colon cancer resection do not completely explain disparities in mortality. Cancer 118(2):469–477.
Roth, C. J., D. A. Clunie, D. J. Vining, S. J. Berkowitz, A. Berlin, J. P. Bissonnette, S. D. Clark, T. C. Cornish, M. Eid, C. M. Gaskin, A. K. Goel, G. C. Jacobs, D. Kwan, D. M. Luviano, M. P. McBee, K. Miller, A. M. Hafiz, C. Obcemea, A. V. Parwani, V. Rotemberg, E. L. Silver, E. S. Storm, J. E. Tcheng, K. S. Thullner, and L. R. Folio. 2021. Multispecialty enterprise imaging workgroup consensus on interactive multimedia reporting current state and road to the future: HIMSS-SIIM collaborative white paper. Journal of Digital Imaging 34(3):495–522.
Ruamviboonsuk, P., R. Tiwari, R. Sayres, V. Nganthavee, K. Hemarat, A. Kongprayoon, R. Raman, B. Levinstein, Y. Liu, M. Schaekermann, R. Lee, S. Virmani, K. Widner, J. Chambers, F. Hersch, L. Peng, and D. R. Webster. 2022. Real-time diabetic retinopathy screening by deep learning in a multisite national screening programme: A prospective interventional cohort study. Lancet Digital Health 4(4):e235–e244.
Salto-Tellez, M., P. Maxwell, and P. Hamilton. 2019. Artificial intelligence—the third revolution in pathology. Histopathology 74(3):372–376.
Sammut, S. J., M. Crispin-Ortuzar, S. F. Chin, E. Provenzano, H. A. Bardwell, W. Ma, W. Cope, A. Dariush, S. J. Dawson, J. E. Abraham, J. Dunn, L. Hiller, J. Thomas, D. A. Cameron, J. M. S. Bartlett, L. Hayward, P. D. Pharoah, F. Markowetz, O. M. Rueda, H. M. Earl, and C. Caldas. 2022. Multi-omic machine learning predictor of breast cancer therapy response. Nature 601(7894):623–629.
Scharpf, R. B., A. Balan, B. Ricciuti, J. Fiksel, C. Cherry, C. Wang, M. L. Lenoue-Newton, H. A. Rizvi, J. R. White, A. S. Baras, J. Anaya, B. V. Landon, M. Majcherska-Agrawal, P. Ghanem, J. Lee, L. Raskin, A. S. Park, H. Tu, H. Hsu, K. C. Arbour, M. M. Awad, G. J. Riely, C. M. Lovly, and V. Anagnostou. 2022. Genomic landscapes and hallmarks of mutant RAS in human cancers. Cancer Research 82(21):4058–4078.
Scherer, A. M., H. O. Witteman, J. Solomon, N. L. Exe, A. Fagerlin, and B. J. ZikmundFisher. 2018. Improving the understanding of test results by substituting (not adding) goal ranges: Web-based between-subjects experiment. Journal of Medical Internet Research 20(10):e11027.
Schnall, M., R. Cruea, and S. E. Seltzer. 2023. The diagnostic cockpit of the future. Journal of the American College of Radiology 20(9):870-874.
Schwartzberg, L. S., G. Li, K. Tolba, A. B. Bourla, K. Schulze, R. Gadgil, A. Fine, K. T. Lofgren, R. P. Graf, G. R. Oxnard, and D. Daniel. 2022. Complementary roles for tissue- and blood-based comprehensive genomic profiling for detection of actionable driver alterations in advanced NSCLC. JTO Clinical and Research Reports 3(9):100386.
Siegel, R. L., A. Jemal, R. C. Wender, T. Gansler, J. Ma, and O. W. Brawley. 2018. An assessment of progress in cancer control. A Cancer Journal for Clinicians 68(5):329–339.
Smyth, L. M., Q. Zhou, B. Nguyen, C. Yu, E. M. Lepisto, M. Arnedos, M. J. Hasset, M. L. Lenoue-Newton, N. Blauvelt, S. Dogan, C. M. Micheel, C. Wathoo, H. Horlings, J. Hudecek, B. E. Gross, R. Kundra, S. M. Sweeney, J. Gao, N. Schultz, A. Zarski, S. M. Gardos, J. Lee, S. Sheffler-Collins, B. H. Park, C. L. Sawyers, F. André, M. Levy, F. Meric-Bernstam, P. L. Bedard, A. Iasonos, D. Schrag, D. M. Hyman, and the AACR Project GENIE Consortium. 2020. Characteristics and outcome of AKT1E17K-mutant breast cancer defined through AACR Project GENIE, a clinicogenomic registry. Cancer Discovery 10(4):526–535.
Takvorian, S. U., E. Balogh, S. Nass, V. L. Valentin, L. Hoffman-Hogg, R. A. Oyer, R. W. Carlson, N. J. Meropol, L. K. Sheldon, and L. N. Shulman. 2022. Developing and sustaining an effective and resilient oncology careforce: Opportunities for action. Journal of the National Cancer Institute 112(7):663–670.
Tao, J., L. Zhu, M. Yakoub, C. Reissfelder, S. Loges, and S. Scholch. 2022. Cell–cell interactions drive metastasis of circulating tumor microemboli. Cancer Research 82(15):2661–2671.
Unger, J. M., D. L. Hershman, D. Martin, R. B. Etzioni, W. E. Barlow, M. LeBlanc, and S. R. Ramsey. 2014. The diffusion of docetaxel in patients with metastatic prostate cancer. Journal of the National Cancer Institute 107(2):dju412.
Vanguri, R. S., J. Luo, A. T. Aukerman, J. V. Egger, C. J. Fong, N. Horvat, A. Pagano, J. A. B. Araujo-Filho, L. Geneslaw, H. Rizvi, R. Sosa, K. M. Boehm, S. R. Yang, F. M. Bodd, K. Ventura, T. J. Hollmann, M. S. Ginsberg, J. Gao, MSK MIND Consortium, M. D. Hellmann, J. L. Sauter, and S. P. Shah. 2022. Multimodal integration of radiology, pathology and genomics for prediction of response to PD-(L)1 blockade in patients with non-small cell lung cancer. Nature Cancer 3(10):1151–1164.
Vázquez-García, I., F. Uhlitz, N. Ceglia, J. L. P. Lim, M. Wu, N. Mohibullah, J. Niyazov, A. E. B. Ruiz, K. M. Boehm, V. Bojilova, C. J. Fong, T. Funnell, D. Grewal, E. Havasov, S. Leung, A. Pasha, D. M. Patel, M. Pourmaleki, N. Rusk, H. Shi, R. Vanguri, M. J. Williams, A. W. Zhang, V. Broach, D. S. Chi, A. Da Cruz Paula, G. J. Gardner, S. H. Kim, M. Lennon, K. Long Roche, Y. Sonoda, O. Zivanovic, R. Kundra, A. Viale, F. N. Derakhshan, L. Geneslaw, S. Issa Bhaloo, A. Maroldi, R. Nunez, F. Pareja, A. Stylianou, M. Vahdatinia, Y. Bykov, R. N. Grisham, Y. L. Liu, Y. Lakhman, I. Nikolovski, D. Kelly, J. Gao, A. Schietinger, T. J. Hollmann, S. F. Bakhoum, R. A. Soslow, L. H. Ellenson, N. R. Abu-Rustum, C. Aghajanian, C. F. Friedman, A. McPherson, B. Weigelt, D. Zamarin, and S. P. Shah. 2022. Ovarian cancer mutational processes drive site-specific immune evasion. Nature 612(7941):778–786.
Vermesh, O., A. Aalipour, T. J. Ge, Y. Saenz, Y. Guo, I. S. Alam, S. M. Park, C. N. Adelson, Y. Mitsutake, J. Vilches-Moure, E. Godoy, M. H. Bachmann, C. C. Ooi, J. K. Lyons, K. Mueller, H. Arami, A. Green, E. I. Solomon, S. X. Wang, and S. S. Gambhir. 2018. An intravascular magnetic wire for the high-throughput retrieval of circulating tumour cells in vivo. Nature Biomedical Engineering 2(9):696–705.
Vermesh, O., A. D’Souza, I. Alam, M. Wardak, T. McLaughlin, F. El Rami, A. Sathirachinda, J. Bell, S. Pitteri, M. James, S. Hori, E. Gross, and S. Gambhir. 2022. Abstract lb560: Engineering genetically-encoded synthetic biomarkers for breath-based cancer detection. Cancer Research 82(12_Supplement):LB560–LB560.
Walsh-Bailey, C., E. Tsai, R. G. Tabak, A. B. Morshed, W. E. Norton, V. R. McKay, R. C. Brownson, and S. Gifford. 2021. A scoping review of de-implementation frameworks and models. Implementation Science 16(1):100.
Yala, A., P. G. Mikhael, F. Strand, G. Lin, K. Smith, Y. L. Wan, L. Lamb, K. Hughes, C. Lehman, and R. Barzilay. 2021. Toward robust mammography-based models for breast cancer risk. Science Translational Medicine 13(578):eaba4373.
Yala, A., P. G. Mikhael, F. Strand, G. Lin, S. Satuluru, T. Kim, I. Banerjee, J. Gichoya, H. Trivedi, C. D. Lehman, K. Hughes, D. J. Sheedy, L. M. Matthis, B. Karunakaran, K. E. Hegarty, S. Sabino, T. B. Silva, M. C. Evangelista, R. F. Caron, B. Souza, E. C. Mauad, T. Patalon, S. Handelman-Gotlib, M. Guindy, and R. Barzilay. 2022a. Multi-institutional validation of a mammography-based breast cancer risk model. Journal of Clinical Oncology 40(16):1732–1740.
Yala, A., P. G. Mikhael, C. Lehman, G. Lin, F. Strand, Y. L. Wan, K. Hughes, S. Satuluru, T. Kim, I. Banerjee, J. Gichoya, H. Trivedi, and R. Barzilay. 2022b. Optimizing risk-based breast cancer screening policies with reinforcement learning. Nature Medicine 28(1):136–143.
Zech, J. R., M. A. Badgeley, M. Liu, A. B. Costa, J. J. Titano, and E. K. Oermann. 2018. Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: A cross-sectional study. PLoS Medicine 15(11):e1002683.
Zehir, A., R. Benayed, R. A. Shah, A. Syed, S. Middha, H. R. Kim, P. Srinivasan, J. Gao, D. Chakravarty, S. M. Devlin, M. D. Hellmann, D. A. Barron, A. M. Schram, M. Hameed, S. Dogan, D. S. Ross, J. F. Hechtman, D. F. DeLair, J. Yao, D. L. Mandelker, D. T. Cheng, R. Chandramohan, A. S. Mohanty, R. N. Ptashkin, G. Jayakumaran, M. Prasad, M. H. Syed, A. B. Rema, Z. Y. Liu, K. Nafa, L. Borsu, J. Sadowska, J. Casanova, R. Bacares, I. J. Kiecka, A. Razumova, J. B. Son, L. Stewart, T. Baldi, K. A. Mullaney, H. Al-Ahmadie, E. Vakiani, A. A. Abeshouse, A. V. Penson, P. Jonsson, N. Camacho, M. T. Chang, H. H. Won, B. E. Gross, R. Kundra, Z. J. Heins, H. W. Chen, S. Phillips, H. Zhang, J. Wang, A. Ochoa, J. Wills, M. Eubank, S. B. Thomas, S. M. Gardos, D. M. Reales, J. Galle, R. Durany, R. Cambria, W. Abida, A. Cercek, D. R. Feldman, M. M. Gounder, A. A. Hakimi, J. J. Harding, G. Iyer, Y. Y. Janjigian, E. J. Jordan, C. M. Kelly, M. A. Lowery, L. G. T. Morris, A. M. Omuro, N. Raj, P. Razavi, A. N. Shoushtari, N. Shukla, T. E. Soumerai, A. M. Varghese, R. Yaeger, J. Coleman, B. Bochner, G. J. Riely, L. B. Saltz, H. I. Scher, P. J. Sabbatini, M. E. Robson, D. S. Klimstra, B. S. Taylor, J. Baselga, N. Schultz, D. M. Hyman, M. E. Arcila, D. B. Solit, M. Ladanyi, and M. F. Berger. 2017. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nature Medicine 23(6):703–713. Erratum in: Nature Medicine 23(8):1004.























































































