Charting a Future for Sequencing RNA and Its Modifications: A New Era for Biology and Medicine (2024)
Chapter: 6 A Bold Vision for the Future of Sequencing RNA and Its Modifications: Conclusions, Recommendations, and a Path Forward
6
A Bold Vision for the Future of Sequencing RNA and Its Modifications: Conclusions, Recommendations, and a Path Forward
The modified RNA isoforms generated from each gene and the associated proteins that write, read, and erase RNA modifications are diverse and complex; they are also critical for various life processes and are important regulators of health and disease (Delaunay, Helm, and Frye, 2023; Flamand, Tegowski, and Meyer, 2023). Nonetheless, efforts to build technologies and conduct research in the field of RNA modifications have thus far been ad hoc and inconsistent. This has impeded a complete understanding of the role of RNA modifications in living systems and has prevented the full realization of RNA-based solutions for issues in the fields of health and medicine, agriculture, and synthetic biology, among others. Chapters 2–5 make the case for the development of technology and surrounding infrastructure that would allow any epitranscriptome to be characterized and interrogated fully. In this chapter, the committee describes key conclusions and recommendations that, if carefully considered and implemented, will propel the field forward, potentially enabling major strides in understanding the basic rules of life, alleviating human suffering, and lessening societal and environmental challenges.
The committee was tasked with examining the scientific need for sequencing RNA modifications. The details of what currently is known about RNA modifications and how this knowledge has been applied to address issues in health and society are detailed in Chapter 2. In brief, on their importance and impacts on health, technology and society, the committee agrees that:
- RNA modifications play fundamental roles in diverse life processes. At the cellular level, nearly every process that depends on gene expression is affected by the numerous types of RNA and their many modifications (Roundtree et al., 2017). At the physiological level, RNA modifications function in a wide variety of processes, including organ development, circadian rhythms, and regulation of metabolism and the immune system (Gilbert and Nachtergaele, 2023; Shi et al., 2020). One key example is the role of RNA modifications in a host’s defense against harmful pathogens. Modifications help to “mark” host cellular RNA as its own, distinguishing it from that of potentially harmful pathogens during an immune response (Leung and Amarasinghe, 2016). Conversely, some pathogens, such as viruses (e.g., SARS-CoV-2, the virus responsible for the COVID-19 pandemic), use
- RNA modifications to alter their RNA genomes and evade a host cell’s defense system (Li et al., 2021). Understanding how viruses and bacteria evade the human immune system is key to developing effective countermeasures for treating viral diseases and antibiotic-resistant bacteria—two of the many compelling reasons to study RNA modifications in living systems.
- RNA modifications have been identified as important regulators of health and disease. These modifications—and the molecular machines that write, read, and erase them—regulate biological processes in healthy living systems. Dysregulation of these processes has been shown to contribute to neurodevelopmental disorders, neurodegenerative diseases, heart disease, autoimmune diseases, cancer, and diabetes (Flamand, Tegowski, and Meyer, 2023; Stojković and Fujimori, 2017; Yanas and Liu, 2019).
-
RNA modifications can be leveraged to address societal problems across sectors such as health, bioindustry, and agriculture. For example, modified RNA molecules can, and are, being used to treat and prevent disease (Winkle et al., 2021), grow synthetic biology applications (de la Torre and Chin, 2021), and address food insecurity (Lowe, 2021; Yu et al., 2021). RNA modifications have been used in RNA-based vaccines and therapies; one prominent example is the messenger RNA (mRNA) vaccine for COVID-19 (Box 2-1), which contains modified pseudouridine to preclude activation of harmful immune responses and increase the efficacy of the vaccine (Barbieri and Kouzarides, 2020; Delaunay, Helm, and Frye, 2023; Mei and Wang, 2023). RNA modifications of various types also show promise in the following areas:
- Vaccines for cancer, using mRNAs, that are in Phase I and II clinical trials (Elkhalifa et al., 2022; Mei and Wang, 2023; Sahin et al., 2020)
- Treating rare diseases, such as spinal muscular atrophy (Winkle et al., 2021)
- Enhancing gene-editing capabilities using guide RNAs (Cox et al., 2017)
- Improving yield and biomass of crops, such as rice and potatoes (Lowe, 2021;Yu et al., 2021)
- Spurring growth in synthetic biology applications and nanotechnology (de la Torre and Chin, 2021; Parsons et al., 2023; Poppleton et al., 2023)
- Efforts to sequence RNA modifications and leverage them for biotechnology and biomanufacturing applications have potential to benefit the U.S. bioeconomy. For instance, efforts directed toward advancing epitranscriptomics could aid in the development of precision multi-omic medicine, which was identified as one of the bold goals for advancing human health, a major sector of the U.S. bioeconomy (White House, 2023). Improvements to RNA-based biotechnology and biomanufacturing would undoubtedly benefit other areas that feed into the bioeconomy, such as bioindustry and agriculture.
- Only in recent years has the science community come to appreciate the importance of RNA modifications in healthy living systems, how their dysfunction can cause disease states, and their potential therapeutic applications, and much remains unknown.
Conclusion 1: RNA modifications are a critical but underexplored area of research. A more complete understanding of RNA modifications will be important for significantly advancing the fundamental knowledge of living systems; maintaining the health of humans, plants, animals, and the environment; preventing and treating disease; improving crop yields and resilience; stimulating the bioeconomy; and addressing other issues of societal importance.
KEY COMPONENTS OF A ROADMAP TO UNLOCK ANY EPITRANSCRIPTOME
The processing and modification of an RNA molecule is amazingly diverse and dynamic. Thus, the collective set of RNA molecules and their modifications, or “epitranscriptome,” differs substantially between every tissue and cell type so that the RNA can meet specific demands—for example, to specify muscle or skin. Further diversity arises from factors such as age, sex, and environment. All of this diversity and plasticity creates orders of magnitude more complexity in sequencing epitranscriptomes than in sequencing the genome. For these reasons, discovering all modifications and their positions in various epitranscriptomes exceeds the challenge presented by the Human Genome Project (HGP). While the HGP aimed to provide complete reference genomes for humans and model organisms, a similar goal for epitranscriptomes would be far more challenging. A more impactful goal is to enable sequencing of any epitranscriptome by developing the necessary technologies and surrounding infrastructure. That said, reference epitranscriptomes that represent specific cell types under defined conditions from both humans and a set of model organisms will be invaluable for comparisons and for setting goals to provide tangible measures of progress. Single-celled prokaryotic or eukaryotic organisms, cultured cells, and well-defined tissues seem ideal for such purposes. Various possibilities for potential reference epitranscriptomes need to be considered carefully and appropriate milestones need to be set.
Conclusion 2: Sequencing the vast array of RNA molecules and discovering all modifications in all of their positions under various conditions and cellular states exceeds the challenge of the HGP. Because there are many important epitranscriptomes to determine, developing technology and infrastructure to enable the determination of any epitranscriptome will be the most impactful goal.
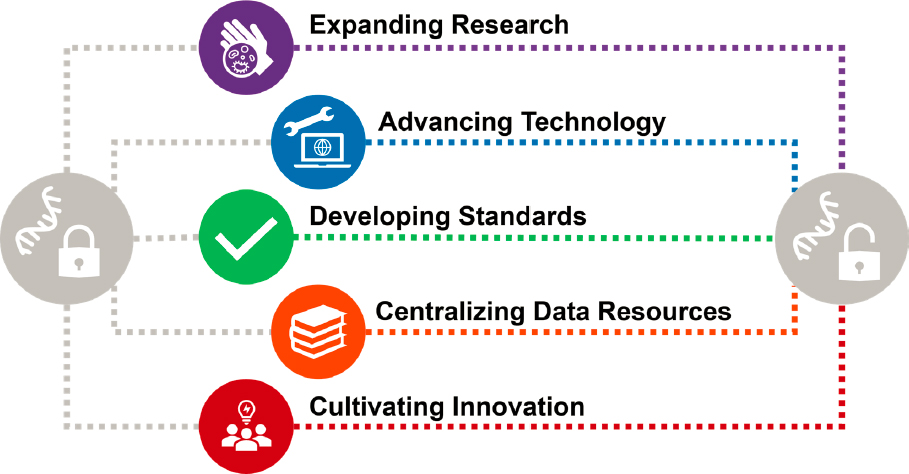
NOTES: Expanding research; advancing experimental and computational tools and technology; developing references and standards of various types; establishing stable, integrated, and centralized data resources; and cultivating innovation across several dimensions, including workforce development, are necessary to fully reveal any epitranscriptome.
The committee advocates for a concerted, large-scale effort around the development of technology and surrounding infrastructure to accelerate the knowledge of RNA modifications and enable the direct end-to-end sequencing of RNA and its modifications. To realize these goals, the committee envisions that multiple efforts will need to occur in parallel, as shown in Figure 6-1.
Expanding Research
Ongoing research in the field of RNA modifications needs to continue and expand. Despite increased recognition of the importance of RNA modifications in health, and their broad application potential for diagnosing, treating, and preventing disease, numerous gaps remain in the understanding of the regulation and function of these modifications. Fundamental research will be critical for identifying the locations of known modifications, discovering additional modifications, and uncovering the functional importance of every modification.
Conclusion 5: Discovery efforts and fundamental research in the field of epitranscriptomics will reinforce the importance and impact of RNA modifications and fuel technological advances that will improve scientists’ ability to sequence them. New funding mechanisms, public and private, that encourage collaboration, spur innovation, and increase interest in RNA modifications will be critical.
Advancing Tools and Technology
Determining the positions of all modifications on individual RNA molecules and elucidating their function in living systems will require significant technological advancement. Currently, there are tools and methods that can identify and map a small subset of the more than 170 RNA modifications, but each suffers from some limitations. Thus, there is a great need in the field to improve the sensitivity, specificity, and throughput of technologies that currently exist and to explore new and emerging instrumentation and methodologies to enhance the capabilities for sequencing RNA modifications and determining their abundance and stoichiometry. A key milestone on the way to reaching this goal is establishing the ability to sequence RNA molecules from end to end, preserving information about all modifications. In Chapter 3, the committee examined the current state of tools and technology and explored possibilities in new and emerging tools that may move the field forward.
On tools and technologies, the committee agrees that:
- Current methods for global measurement of RNA modifications cannot identify or quantify all modifications in complex mixtures at high accuracy and with high precision.
- There is a need to better define and standardize appropriate sample isolation, preparation, and handling methods for global analyses. Several recent reports document how sample handling and preparation steps prior to global analysis by LC-MS/MS can lead to artifacts that are incorrectly reported as new modifications. These processes can also alter the chemical structure of an existing modification.
- Methods for cataloging and quantifying RNA modifications, even without positional information, will be important for facilitating the ultimate technological goal. Although it cannot predict what advances will enable end-to-end sequencing of RNA and all of its modifications, or when that might occur at the current level of investment, the committee is confident that efforts to improve upon existing methods, such as those available for cataloging and quantifying RNA modifications, even without knowing their sequence context, will be an important initial step toward reaching this goal.
- Such advances would enable discovery of new modifications and may drive and inform development of new sequencing technologies. To support these methods, improvements in instrumentation, sensitivity, and the availability of standards will be needed.
- Indirect sequencing methods are high throughput and low cost, and importantly, provide positional information. However, they do not always provide information on the entire length of RNAs or enable discrimination of individual RNA molecules. This poses significant challenges for interrogating all modifications on a single RNA. Furthermore, to enable single-cell analyses, the sensitivity and specificity of indirect sequencing methods need to be improved.
- Direct sequencing methods also provide positional information and hold the greatest promise for end-to-end sequencing of RNA and all of its modifications; however, these methods currently are in early stages of development, are expensive, require large sample input, and are prone to errors.
- While computational methods used with indirect sequencing perform well, advances in computational methods for direct sequencing and mass spectrometry are ripe for improvement. Signal data extraction, basecalling, quality control, and sequence assembly and alignment as part of direct sequencing workflows all need improved computational methods to increase accuracy, extend reads, expand the pool of detectable modifications, and reduce errors. Mass spectrometric workflows need more and better software packages that are tailored for mapping RNA modifications against an array of genomic and transcript data.
- Dedicated approaches specific to RNA and its unique properties are needed. Tools, technologies, and methods for extraction, analysis, and sequencing of RNA modifications significantly lag behind those available for studying DNA and proteins. New approaches and technologies are needed that can account for the diversity of RNA isoforms, the variable expression level of RNAs giving rise to variable amounts of material, the dynamic nature of RNA structures and chemical modifications, the interplay of RNA structure and chemical modification, and how all of these factors are influenced by the location of RNA within the cell. Protocols designed to maintain the integrity of RNA and optimized for specific downstream analyses would accelerate progress and allow direct comparisons of data from different sources.
- Future technological and methodological innovations need to offer turn-key solutions for use by a broader research community than is currently possible. Workflows using currently available technologies and methods are cumbersome and require highly skilled users who are trained across multiple disciplines; thus, these technologies and methods are inaccessible or impractical for smaller labs and researchers who do not study RNA exclusively.
Conclusion 6: The current tools, technologies, and methodologies for end-to-end sequencing of RNA and all of its modifications are insufficient. The field of RNA biology will be driven forward by improving upon existing approaches and advancing new technologies that are robust and quantitative, and that preserve the information of full-length RNAs.
Conclusion 7: Improving the sensitivity of methodologies for cataloging and quantifying all RNA modifications in a sample, even without positional information, is an important enabling step that will inform the development of future RNA sequencing technologies and facilitate discovery of additional RNA modifications. Achieving this crucial intermediate goal will be spurred by an expanded repertoire of modified nucleosides for use as reference standards and more sensitive instrumentation.
Conclusion 8: Efforts directed toward enabling end-to-end sequencing of RNA and its modifications will accelerate innovation in the life sciences research enterprise but will also pave the way for developing new biotechnologies (e.g., biotherapeutics, vaccines, diagnostics, nanomaterials) and novel approaches that open new doors in life sciences research and other areas that are not yet apparent.
Public and private investment and partnership will be important for reaching these technological goals. The National Institutes of Health (NIH) is already invested in building the technology and capabilities to study and sequence RNA and their modifications. Specifically, the National Human Genome Research Institute (NHGRI) and the National Institute of Environmental Health Sciences (NIEHS) have demonstrated leadership in addressing the scientific and technological gaps for sequencing RNA modifications and the committee for this report has built upon this important foundation.1 NHGRI already funds two research centers focused on studying epitranscriptomes,2 and NIEHS recently launched the Functional RNA Modifications Environment and Disease program, an initiative to investigate the impact of environmental exposures on RNA modifications.3 The National Science Foundation (NSF) and several other institutes and centers at NIH—such as the National Institute of General Medical Sciences and the National Institute of Allergy and Infectious Diseases—also fund research on RNA modifications.4 In terms of private foundations, The Warren Alpert Foundation has significantly invested in RNA modifications research and technology and infrastructure development to improve human health.
Other U.S. federal entities—such as the Defense Advanced Research Projects Agency (DARPA), Advanced Research Projects Agency for Health (ARPA-H), and Department of Energy (DOE)—could invest in advancing capabilities to gain knowledge about RNA modifications through new and improved measurement methods and sequencing technologies; doing so would directly benefit the ability to leverage such knowledge for biotechnology and manufacturing applications for health and medicine and other major sectors—a stated priority for the U.S. government. Additionally, given that advances in computational methods are critical to achieving the goals set out in this report, specific calls for funding to support collaborative initiatives that integrate experimental and computational research and development must be prioritized. The National Institute of Standards and Technology (NIST) will undoubtedly play an important role in developing and curating standards to support tool and technology development.
Recommendation 2: Federal funders of research—such as the National Institutes of Health, National Science Foundation, Department of Defense, and Department of Energy—should invest in and prioritize (a) addressing limitations and closing gaps in the existing tools and technologies available for epitranscriptomics, (b) exploring new and emerging approaches, and (c) compiling and centralizing resources pertaining to available tools and methods. Existing tools and technologies should be refined and optimized, and novel approaches to characterize RNA modifications should be explored, with the goal of enabling end-to-end sequencing of RNA and its modifications. Information about
___________________
1 In 2022, NHGRI and NIEHS collaborated to host a virtual workshop “to determine the current capabilities, needs, and prospects for comprehensive characterization and understanding of the true diversity of all RNAs and their modifications at a chemical and structural level in relation to normal and disease states.” See https://www.niehs.nih.gov/news/events/pastmtg/2022/rnaworkshop2022/index.cfm (accessed November 15, 2023).
2 See https://www.genome.gov/Funded-Programs-Projects/Centers-of-Excellence-in-Genomic-Science/CEGS-Awards (accessed November 14, 2023).
3 See https://factor.niehs.nih.gov/2018/7/science-highlights/council (accessed November 15, 2023).
4 NIH RePORTER showing projects relating to RNA modifications by NIH institutes and centers https://reporter.nih.gov/search/MzH7kj-gskaGeTY-6QHl3g/projects/charts (accessed November 15, 2023).
available experimental methods and associated computational approaches should be compiled and centralized as a resource that researchers can use to understand the utility, biases, strengths, and weaknesses of different methods and tools. It will be critical to use a diversity of funding mechanisms and models, and to encourage and support collaborative initiatives that integrate experimental and computational components.
With a concerted investment of time, effort, and funding by key public and private groups, the committee hopes that within 15 years, sensitive and specific technologies, methods, and computational tools will be developed that are capable of identifying RNA modifications, and determining their location and abundance on an RNA molecule in a single experiment—and eventually, at the single-cell level (Figure 6-2). Such tools would make it possible to interrogate whole epitranscriptomes with resolution at the level of individual RNA isoforms and their modifications, revealing unprecedented insights into the influence of RNA modifications on the folding, stability, and function of a single RNA molecule. Early, accessible targets for developing new tools and technologies include small and high-abundance RNAs, such as transfer RNAs (tRNAs). Likewise, epitranscriptomes of several high-importance viral pathogens would be a valuable and practical payoff for the first phase of this effort. Soon after, complete epitranscriptomes for human cultured cells and tissues may be in reach, and eventually multicellular eukaryotes. Enhanced capabilities will one day allow researchers to study clinical samples to understand disease and generate personalized treatments.
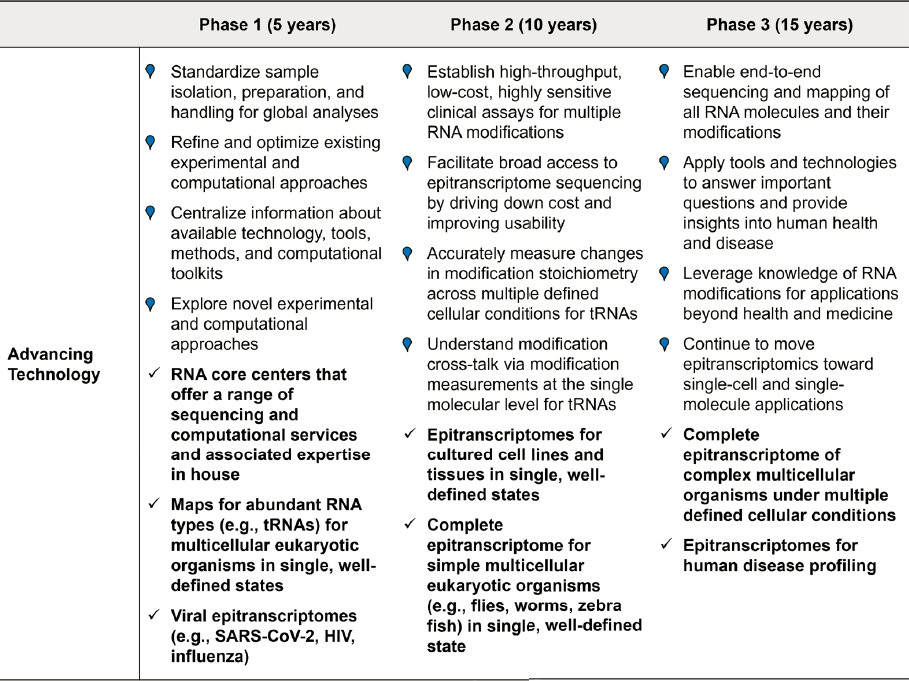
Roadmap for Advancing Tools and Technologies
Within 5 years:
- Standardize sample isolation, preparation, and handling methods for global analyses.
- Establish RNA core centers that offer a range of expertise, techniques, and methods spanning the sequencing approaches (e.g., global modification mapping, direct and indirect sequencing), as well as the computational and bioinformatics expertise needed to analyze and interpret the data.
- Refine and optimize existing approaches.
- Improve the sensitivity and specificity of instrumentation used for cataloging and quantifying all RNA modifications in a single sample. Develop new enzymes and chemicals to facilitate improved sensitivity and specificity of modification detection.
- Refine experimental and corresponding computational approaches for indirect and direct sequencing, such as basecalling algorithms, that can simultaneously determine the presence and quantity of multiple RNA modifications with high accuracy and precision.
- Begin research to extend these capabilities to the single-cell level, which will enable dynamic understanding of specific modifications without averaging over different cell types or cellular states. Improving sensitivity of methods and compatibility of workflows will be important for accomplishing this.
- Compile and centralize existing information about available technology, tools, and methods and associated computational toolkits.
- Compile resources for informing users about the strengths, weaknesses, and potential biases of different methods.
- Centralize the databases and make software tools easily accessible for both experienced and first-time epitranscriptomics researchers and clinicians.
-
Explore novel approaches.
- Promote the development of new ideas and new technologies for sequencing RNA modifications by providing forums and opportunities for innovative, cross-disciplinary thinking.
- Expand opportunities for collaborations and partnerships among academic, industrial, and government sectors.
- Encourage innovation in research by investing in novel approaches for epitranscriptomics.
- Map modifications for abundant RNAs, such as tRNAs and rRNAs, for multicellular eukaryotic organisms in single, well-defined states, including stoichiometry.
- Determine epitranscriptomes for viral pathogens, such as SARS-CoV-2, HIV, and influenza.
Within 10 years:
- Establish high-throughput, low-cost, highly sensitive clinical assays for multiple RNA modifications relevant to human health and disease.
- Facilitate broad access to epitranscriptome sequencing by driving down costs and improving usability.
- Accurately measure changes in modification stoichiometry across multiple defined cellular conditions for RNAs with high modification diversity (e.g., tRNAs).
- Understand modification crosstalk via modification measurements at the single-molecule level for RNAs with high modification diversity (e.g., tRNAs).
- Characterize epitranscriptomes of cultured cell lines and tissues in single, well-defined states.
- Complete the first epitranscriptome map of one or more simple multicellular eukaryotic organisms in a single, well-defined state.
Within 15 years:
- Enable end-to-end sequencing and mapping of all RNAs and their modifications.
- Apply tools and technologies for answering important scientific questions and providing insights into human health and disease.
- Leverage knowledge of RNA modifications for applications beyond health and medicine in other key areas, such as agriculture and synthetic biology.
- Continue to move epitranscriptomics toward single-cell and single-molecule applications.
- Complete epitranscriptomes of multiple, complex multicellular eukaryotic organisms under multiple defined cellular conditions.
- Profile RNA modifications in human diseases.
Developing Standards
Several types of standards are needed to support research and technology development for the RNA modifications field. In Chapter 4, the committee examined the current state of standards and needs for addressing major gaps and challenges.
On standards, the committee agrees that:
- A broad collection of modified RNA reference materials is needed to support efforts in fundamental research and tool and technology development. Such reference materials need to be technology agnostic and include modified nucleosides and oligonucleotides of known sequence and modification stoichiometry. These reference standards would support
NOTE: tRNA = transfer RNA.
- technical validation of methods and assays and cross-referencing of results obtained using different approaches or by different laboratories.
- Federal involvement and leadership are needed to support the creation of modified RNA reference materials at scale. Although synthetic routes for generating modified nucleosides exist in many cases, limited commercial or nonprofit sources for these standards exist. While the committee heard from several for-profit enterprises on their interest and continued investment in this area, the lack of broad-based market forces supporting the creation of such materials at scale emphasizes the need for federal involvement and leadership, as well as public–private partnerships.
- Systematic nomenclature is needed for representing a modified nucleoside within an overall RNA sequence. Clear and consistent guidance, and standards for modified RNA nomenclature, and standards for representation of modified RNA datasets are needed to simplify data sharing and access—these would accelerate open access and sharing of information about RNA and its modifications.
- Clear guidance on data quality is needed to facilitate appropriate deposition of data to a centralized repository. Establishing such guidance and its enforcement will enhance scientific reproducibility, rigor, and reuse of datasets and will accord with the open science principles promoted in the United States and internationally (UNESCO, 2023; Nelson, 2022).
- Progress in creating data and database standards will lead to the creation of more widely accessible and informative databases, once such information can be shared more easily among database platforms.
- NIST is well positioned to lead the development, curation, and promotion of standards for epitranscriptomics. This effort for epitranscriptomics falls directly within NIST’s mission to “promote U.S. innovation and industrial competitiveness by advancing measurements in science, standards, and technology toward economic security and improved quality of life” (“NIST General Information”, 2008p. 2).5 The Biosystems and Biomaterials Division6 at NIST may be an appropriate group to lead the development of RNA reference materials.
- The National Center for Biotechnology Information (NCBI), a division of the National Library of Medicine at NIH, “develops and promotes standards for databases, data deposition and exchange, and biological nomenclature” (“Our Mission”, n.d., p. 4) as a part of its responsibilities, and could take on the role of establishing and promoting data and database standards for epitranscriptomics.
Conclusion 9: Several types of standards are needed, specifically (a) technology-agnostic modified RNA reference materials that enable assay validation and cross-referencing of approaches, (b) data standards around nomenclature and clear guidelines for data deposition and exchange, and (c) robust and sustainable platforms for the curation and indexing of vast amounts of RNA data.
Recommendation 3: The National Institute of Standards and Technology should develop, curate, and promote standards to support the field of epitranscriptomics. Specifically, modified RNA reference materials should be developed with a focus on making them widely available and affordable.
Recommendation 4: The National Center for Biotechnology Information should establish and promote standards for databases, data deposition and exchange, and nomenclature for RNA modifications.
If the above actions are taken, the committee envisions that within 15 years, custom-order, affordable oligonucleotides of any sequence, length, modification stoichiometry, and structure could be readily available for use in research and technology development. With the establishment of data and database standards, it could also be possible to have seamless access to universally available and shareable information on RNAs of any biotype from any defined set of cellular conditions, including their modification status and their biological, medical, and functional properties. The committee developed milestones and deliverable to achieving this vision (Figure 6-3).
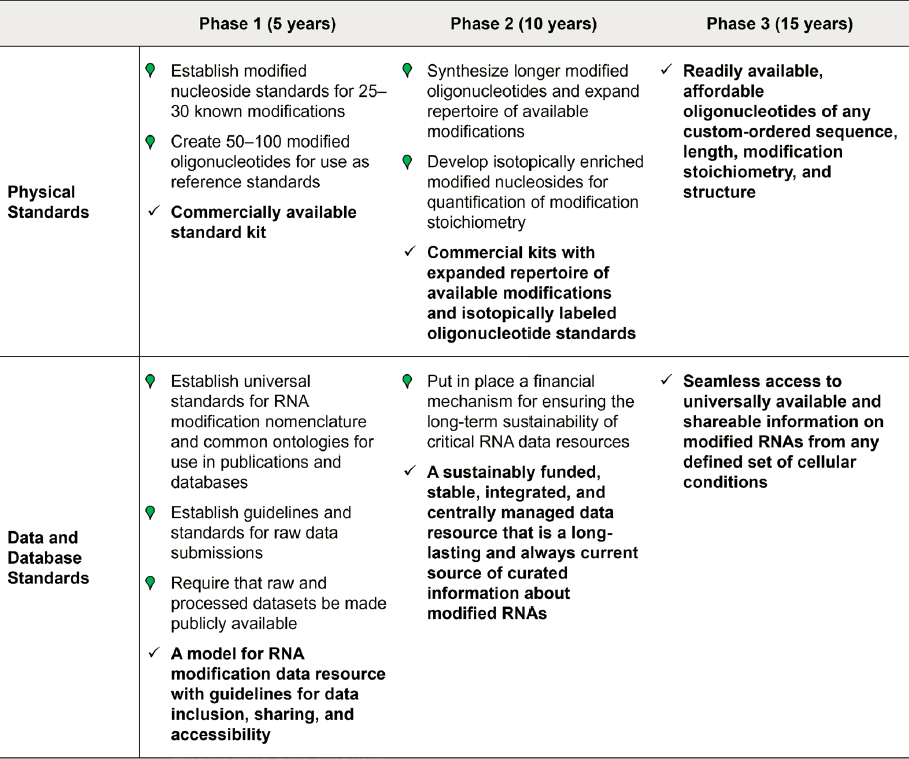
Roadmap for Developing Standards and Databases
Within 5 years:
-
Physical standards:
- Establish modified nucleoside standards for 25–30 known modifications of biological significance and ensure that these are available to be used at scale.
- Create a suite of 50–100 modified oligonucleotides of known sequence and known
___________________
5 See https://www.nist.gov/director/pao/nist-general-information (accessed November 14, 2023).
6 See https://www.nist.gov/mml/bbd/about-bbd (accessed November 14, 2023).
-
- modification stoichiometry for use as standards for evaluating the technical accuracy and reproducibility of instrumentation.
- Develop a commercially available standard kit that includes a suite of modified reference standards for nucleosides and oligonucleotides.
-
Data standards:
- Establish universal standards for RNA modification nomenclature and common ontologies for use in publications and databases.
- Establish standards and guidelines for submission of raw data to journals.
- Require that raw signal data and processed datasets be made publicly available through centrally managed databases.
-
Database standards:
- Collaborate with key public, private, and international groups to define standards necessary for developing and managing RNA modification databases and resources.
- Establish an agreed-upon model for RNA modification data resources with guidelines for data inclusion, sharing, and accessibility.
Within 10 years:
-
Physical standards:
- Partner with companies to synthesize longer modified oligonucleotides and expand repertoire of available modifications.
- Develop isotopically enriched versions of modified nucleosides for enhanced quantification of modification stoichiometry.
- Develop commercial kits with an expanded repertoire of available modifications and isotopically labeled oligonucleotide standards.
-
Database standards:
- Put in place a financial mechanism for ensuring the long-term sustainability of critical RNA databases.
- Promote a sustainably funded, stable, integrated, and centrally managed data resource that is a long-lasting and always-current source of curated information about modified RNAs.
Within 15 years:
- Readily available and affordable oligonucleotides of any custom-ordered sequence, length, modification stoichiometry, and structure.
- Seamless access to universally available and shareable information on all RNAs from any defined set of cellular conditions, their modification status, and their appropriate biological, medical, or other functional aspects. Ensure that repositories of this information are financially viable.
Centralizing Data Resources
Easy access to reliable, accurate, and up-to-date information about RNA modifications is key to advancing epitranscriptomics. In Chapter 4, the committee described current databases that exist, and major challenges and gaps that need to be addressed in this space.
On databases, the committee agrees that:
- There are several current databases that store information on modified RNAs (see Table 4.1). However, these databases are largely managed by individual research laboratories, may have a narrow focus on a particular modification, or curate information based only on a specific type of RNA.
- While these RNA databases have been vital to advancing the field of RNA biology, a major concern is the loss of resources (e.g., funding, staff) leading to a lack of maintenance or abandonment of carefully curated databases, which poses the risk of limiting scientific growth and understanding while wasting time, effort, and resources.
- Centralized administration of RNA data resources is needed to ensure sustainable funding and stability, so that the current knowledge on RNA modifications can be integrated and new modifications can be indexed. NIH is well-suited to establish, maintain, and coordinate databases for RNA modifications data. Specifically, NCBI is an established national resource, and housing epitranscriptomic data falls squarely within its mission7 to develop new information technologies to aid in the understanding of health and disease. Within NCBI, the Information Engineering Branch,8 may assist with building
___________________
7 NCBI Mission https://www.ncbi.nlm.nih.gov/home/about/mission/ (accessed November 15, 2023).
8 Organizational structure of NCBI https://www.ncbi.nlm.nih.gov/home/about/structure/ (accessed November 15, 2023).
- databases, integrating data resources, and developing and promoting standards for such resources.
Conclusion 10: The prevalence of “home-grown,” small-group-supported RNA databases has been vital to advancing the field of RNA biology. Nonetheless, a major concern is the loss of resources (e.g., funding, staff) leading to a lack of maintenance of these laboratory-housed databases. Abandoning carefully curated databases may limit scientific growth and understanding, and waste time, effort, and resources.
Recommendation 5: The National Institutes of Health (NIH) should establish and maintain a sustainably funded, stable, integrated, and centrally managed database (or ensemble of databases) that is a long-lasting and always-current source of curated information about RNAs and their modifications. Such a resource could build upon, through mirroring or linkage, existing well-maintained databases that contain valuable information related to RNA modifications. Efforts to develop such centralized databases should strive to provide accurate, single-molecule, end-to-end information on RNA modifications. NIH should initiate U.S. collaboration with other countries invested in research on RNA and its modifications. In consultation or partnership with the National Institute of Standards and Technology, standards for the deposition and exchange of experimental raw data should be developed and promoted according to FAIR (findability, accessibility, interoperability, and reusability) principles to ensure data in the field of RNA modifications are accessible, well maintained, and user friendly.
Cultivating the Future Workforce
Critical to advancing the field of epitranscriptomics is support for a well-informed, well-trained, and diverse workforce, comprising both retrained professionals and the next generation of scientists. Chapter 5 describes the current needs for the education and training of a workforce specific to the RNA modifications field.
On education, training, and workforce development, the committee agrees that:
- The study of RNA and its modifications is an increasingly interdisciplinary endeavor, and training programs for the RNA modifications workforce need to reflect this. Disciplines such as RNA biology, molecular biology, biochemistry, chemistry, engineering, physics, and computer science need to be integrated into learning modules, workshops, and training programs for RNA biologists, specifically, and life sciences, more generally. In undergraduate education, the chemistry (e.g., structure and properties) of RNA modifications and the biology (e.g., regulation and function) of modified RNAs are topics that need to be integrated into general molecular biology and biochemistry courses. Advanced courses in RNA biology can also be developed and made available to stimulate a more specialized focus. Computational competence has become an essential skill for today’s STEM (science, technology, engineering, and mathematics) workforce, given the need to analyze ever-increasing amounts of high-throughput data, and the field of epitranscriptomics is no different. Therefore, biology courses need to also provide training in computer science, computational biology, and bioinformatics.
- To recruit and retain students, it is important to ensure that they gain both an awareness of RNA biology as an important and exciting field and knowledge of technologies that are emerging from the field. Promoting engagement by sharing information about relevant advances, such as the success of mRNA vaccines, can attract students from
- diverse backgrounds who are interested in careers in STEM and careers with positive societal impacts.
- The RNA modifications field is growing rapidly; attracting more individuals with diverse expertise, experience, and interests is critical for the current and future success of the epitranscriptomics community. Opportunities and mechanisms for convening diverse groups could have a transformative impact on solving the technological challenges associated with sequencing all RNAs with their full collection of modifications.
- Private and public funding agencies will play an important role in supporting the development of additional educational and learning materials. In particular, NSF can lead in this space because of their specific responsibility and mission for investing in research to expand knowledge in science, engineering, and education.9 Education and pedagogy experts, along with scientific societies with ties to the RNA research and broader life sciences communities, will also be important players in the education of future scientists who work in the field of RNA modifications.
- Individuals already in the workforce can make important contributions to achieving the goals described above and to the application of epitranscriptomics. These individuals need to be considered intentionally during hiring and recruitment, and offered professional development, retraining, and continuing education opportunities tailored to their needs—either from in-house workforce development programs (in the case of large companies) or through community-based intermediary organizations, local colleges and universities, and professional organizations. Human resources personnel, hiring managers, executives, and other individuals who review applicants can expand the pool of hires by staying up to date on technical terminology and seeking out various pathways both within and outside of academic degree programs where individuals have built skills and expertise translatable to jobs in RNA biology. Organizing workshops, forums, panels, and technical courses held at RNA and biochemistry conferences will allow more people to learn about RNA biology outside of a classroom setting.
Conclusion 11: Greater emphasis on RNA science in undergraduate courses is needed to build a better infrastructure for embracing future generations in the workforce. In addition to further education, the existing and future workforce needs interdisciplinary training with strong quantitative and computational skills.
Conclusion 12: Educational efforts in the RNA modifications field need to (a) use methods that promote engagement, (b) reflect the interdisciplinary nature of the science in education and related workforce development efforts, (c) invest in reaching and engaging students and trainees from diverse backgrounds, and (d) scale up proven strategies for retaining trainees in piloted programs.
Recommendation 6: Institutes and funding agencies, such as the Howard Hughes Medical Institute, National Institute of General Medical Sciences, and National Science Foundation—in consultation or partnership with relevant education and pedagogy experts; scientific societies, such as the RNA Society, American Chemical Society, and American Society for Biochemistry and Molecular Biology; and industry groups, such as the Parenteral Drug Association and the International Society for Pharmaceutical Engineering—should build upon existing educational materials and training opportunities for high school, undergraduate, graduate, and postgraduate groups, and for the
___________________
9 See NSF’s Education and Training page https://new.nsf.gov/focus-areas/education (accessed December 19, 2023).
private sector. Such materials and opportunities should be tailored to fit the needs and interests of each group and should cover the basic biological, chemical, and biochemical principles of RNA modifications and the tools available for their study. All materials should incorporate engaging examples that demonstrate the importance of RNA and its modifications in fundamental science, health and medicine, food safety, the environment, and manufacturing.
The committee envisions that with proper attention to education, training, recruitment, and retention, a well-trained, impassioned, diverse U.S. workforce with interdisciplinary expertise will be able to apply and advance sophisticated RNA biology, including epitranscriptomics, across the public and private sectors, academia, and industry. This roadmap is designed to aid in the development of education and training resources to build the future workforce for the RNA modifications field in the next 15 years (Figure 6-4).
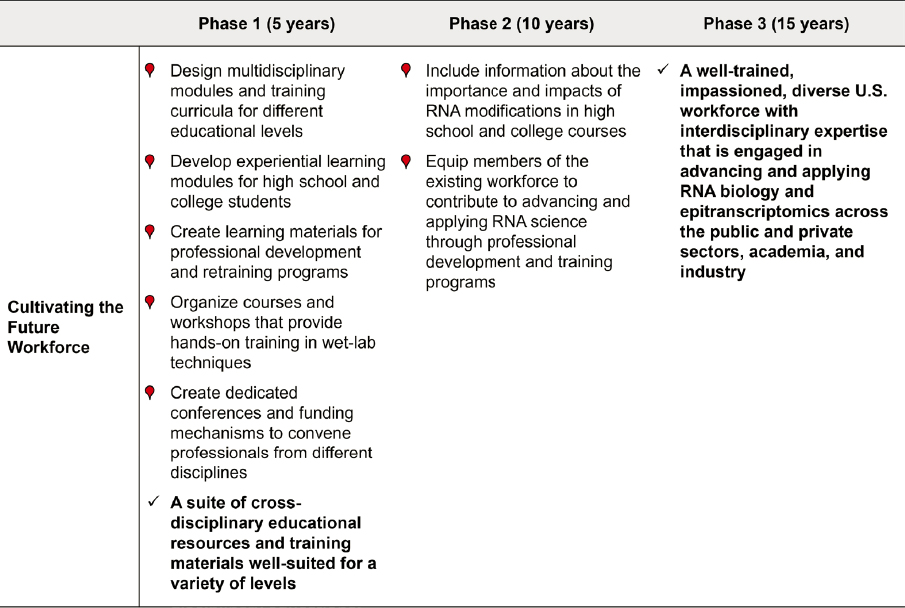
Roadmap for Education, Training, and Workforce Development
Within 5 years:
- Design multidisciplinary modules and training curricula for use by educators at different levels with materials designed such that they can be easily incorporated into current curricula.
- Expand curricular materials that integrate open-ended and real-life examples of relevance, as well as hands-on activities and learning modules for high school and college students.
- Develop materials tailored to adult learners for incorporation in professional development and retraining programs offered by private- and public-sector organizations.
- Organize courses and workshops that provide hands-on training in wet-lab techniques and in the analytical framework for sequencing RNA modifications. These workshops would cover the state of the art and accepted standards in the field of RNA modifications and sequencing.
- Create dedicated conferences and funding mechanisms to convene professionals from different disciplines relevant to the RNA modifications field.
- Develop a suite of cross-disciplinary educational resources and training materials for high school, college, and graduate-level educators that incorporate hands-on, real-life examples for use in training the future workforce and public.
Within 10 years:
- Include information about RNA modifications in high school and college courses.
- Equip members of the existing workforce to contribute to advancing and applying RNA science through widely available and accessible professional development and training programs.
Within 15 years:
- Develop a well-trained, impassioned, diverse U.S. workforce with interdisciplinary expertise that is engaged in advancing and applying sophisticated RNA biology, including epitranscriptomics, across the public and private sectors, academia, and industry.
While the components listed above will help drive innovation, other factors are essential for transforming the field of RNA modifications. These other drivers include centralized facilities to perform specialized tasks, readily available, high-quality reagents and research materials, organization and coordination at the national and international levels, and a supportive policy environment
(discussed in Chapter 5). To achieve the improvement of existing technologies and the exploration of new, innovative options for sequencing RNA and its modifications; developing standards to support and drive technology and database development; establishing well-maintained, long-term-funded, and accessible databases; and supporting and sustaining a well-informed workforce; the committee also highlights the need for:
- The engagement of industry and academia in technology and tool development through competitive and collaborative mechanisms.
- Public- and private-sector funding in multiple forms at various scales (e.g., cooperative agreements and investigator-initiated projects, programs reaching large, established and small, start-up laboratories and companies).
- An end market sufficient to attract companies and drive innovation.
- A strategy that recognizes the benefits of international collaboration while maintaining global leadership.
LAUNCHING A LARGE-SCALE INITIATIVE
To support and coordinate the efforts required to unlock any epitranscriptome, a concerted, large-scale initiative is needed to promote and resource the development of technology and associated infrastructure. A major lesson learned from other successful large-scale initiatives, most notably the HGP, is that focused and concerted organization and funding directed toward a common
goal accelerates technological innovation. Inspired by the success of the HGP, the United States has led several other large-scale, coordinated efforts in the life sciences, such as the Glycoscience Program, the Human Microbiome Project, and the Brain Research through Advancing Innovative Neurotechnologies (BRAIN) Initiative.
Conclusion 3: Large-scale, coordinated efforts in the life sciences, such as the HGP, the Glycoscience Program, the Human Microbiome Project, and the BRAIN Initiative, have proven vital in driving innovation in science and technology. Such efforts hold value in their ability to align federal agencies; support public–private partnerships; organize consortia; fund individual laboratories; and prioritize closing gaps in the areas of technology development, synthesis of standards, infrastructure buildout, workforce training, and public awareness.
In these efforts, industry, philanthropic organizations, research institutions, and government funders all contributed to achieving the goal of dramatically accelerating progress in a high-priority area of research. U.S. government research funding sources such as NIH, NSF, DOE, and Department of Defense are major drivers of technological innovation. However, more funding opportunities are needed from these agencies to promote the critical early-stage development of technology toward the goal of end-to-end sequencing of RNA and its modifications. Such opportunities, such as the Small Business Innovation Research and Small Business Technology Transfer programs, support small businesses and academic research laboratories. Other potential loci for government funding include the DARPA Biological Technologies Office, under DOD, and the new ARPA-H, under NIH, because of their focus on research problems of great practical importance and high-risk, high-impact projects. In addition, funding from private foundations is essential for supporting technology development and driving innovation. For instance, the Warren Alpert Foundation, a sponsor of this study, recently funded research and technology development related to RNA modifications.10 The Margot and Thomas Pritzker Family Foundation has also committed funds toward establishing a plant biology center that includes research on how RNA modifications may impact plant growth and crop yields.11 Awards that support cooperative research efforts (e.g., multiple academic labs or academic–industry partnerships) will provide important opportunities for accelerated research progress.
Interfaces that connect the international community of scientists and funding agencies are needed to identify areas for collaboration and facilitation of the sharing of information that may be critical to the overall well-being of humanity and the planet. U.S. government agencies have made several recent calls for research proposals or announcements of awards that relate to RNA research. A few pertinent examples include:
- Partnerships to Transform Emerging Industries—RNA Tools/Biotechnology,12 a recent program solicitation by NSF in coordination with the NHGRI, as part of NSF’s Molecular Foundations for Biotechnology. This program calls for “creative, cross-disciplinary research and technology development proposals to accelerate understanding of RNA function in complex biological systems and to harness RNA research to advance biotechnology” (National Science Foundation, 2023, p. 2).
- Curing the Uncurable via RNA-Encoded Immunogene Tuning (CUREIT).13 In August 2023, ARPA-H announced up to $24 million in funding for the CUREIT project.
___________________
10 See https://www.warrenalpertfoundation.org/home/prior-grantees/ (accessed November 13, 2023).
11 See https://news.uchicago.edu/story/uchicago-receives-10-million-new-pritzker-plant-biology-center (accessed January 4, 2024).
12 See https://www.nsf.gov/pubs/2023/nsf23554/nsf23554.htm (accessed November 14, 2023).
13 See https://arpa-h.gov/news/first-baa/ (accessed November 14, 2023).
- The aim of this effort is to train the immune system to fight cancer and other diseases more effectively through the development of generalizable mRNA platforms.
The international landscape of RNA-focused efforts presents both direct competition and opportunities for development in RNA-based knowledge and tools among U.S. laboratories and industries; U.S. global leadership in this space is by no means assured. Other countries already have made major investments in RNA research. Germany is an international leader with a large-scale effort focused on RNA modifications. Australia and Canada are making major investments in this space, with efforts focused on RNA chemistry and RNA-based therapeutics. To ensure U.S. global leadership in this space, proactive identification of areas for international cooperation and a significant whole-of-government investment of time, funding, and other resources will be needed.
The committee examined other scientific initiatives as it deliberated about the framework needed to integrate and coordinate the efforts toward sequencing RNA and all of its modifications. DNA sequencing technologies were driven forward by the large-scale coordinated effort to map and sequence the human genome. Coordination among the diverse, multidisciplinary groups with vested interests and roles to play in the field of epitranscriptomics will be needed to advance a large-scale initiative in epitranscriptomics in a directed and strategic fashion. NIH has led most of the efforts around RNA modifications research focused on human health and disease. However, the committee feels that a large-scale, coordinated scientific effort focused on the cross-cutting field of epitranscriptomics would have sweeping impacts across various sectors beyond health and fundamental research. Advances in the field of epitranscriptomics likely will impact capabilities in food and agriculture, synthetic biology, nanotechnology, and national security and defense. Given this, a whole-of-government investment in a large-scale epitranscriptomics initiative would align with the missions of the U.S. Department of Agriculture, DOE, DOD, and other groups housed under the U.S. Department of Health and Human Services (such as the Food and Drug Administration and ARPA-H) or U.S. Department of Commerce (e.g., NIST and the National Institute for Innovation in Manufacturing Biopharmaceuticals).
Rather than predetermining a lead agency or specifying a single entity to lead this large-scale scientific effort, the committee feels strongly that key federal officials, program officers, and researchers and contributors from academia and industry and other vested parties need to form a national research consortium. Establishing a U.S. consortium for RNA modifications will be instrumental for accelerating the pace of technology development and for establishing standards, unified protocols, and public databases to facilitate data dissemination and resource sharing among the research community. The resources generated through such a consortium could be readily integrated with datasets produced from other relevant large-scale efforts, such as the ENCODE (Encyclopedia of DNA Elements) project, which has established standards and datasets related to RNA–protein interactions in cells.
Conclusion 4: A large-scale effort focused on epitranscriptomics is needed to accelerate technological innovation and scientific progress in the field. Such an effort will require expertise spanning multiple scientific disciplines (e.g., engineering, computer science, life science, social science) and will impact several sectors (e.g., health, agriculture). An endeavor of this scale and scope will entail a substantial investment of time and resources. Dedicated funding to key federal entities—such as NSF, NIH, NIST, DOD, and the DOE—is critical to enhance their ability to work with academia, industry, philanthropic organizations, and international partners in driving innovation towards sequencing RNA and its modifications and ensuring translation of the resulting scientific breakthroughs into advancements including new, effective biotechnology products.
Recommendation 1: An established oversight body, such as the Office of Science and Technology Policy or a similar entity with appropriate breadth and authority, should catalyze and coordinate efforts supporting a large-scale epitranscriptomics initiative to ensure effective use of resources and minimize duplication. Expertise from the health, agriculture, commerce, energy, national security, and defense sectors will be required. Both research and regulatory agencies should be included as a part of the effort. An implementation plan should be developed and include support for agencies to work with partners in academia, industry, scientific societies, private foundations, international partners, and other relevant groups. The coordinating body should be responsible for strategic coordination of government, academic, and industry partners. The implementation plan should embrace conclusions and recommendations from the committee and aim to do the following:
- Support and fund ongoing and future research and development efforts.
- Close gaps in tools and technology through diverse funding mechanisms that encourage and support integrated experimental and computational collaborations and initiatives.
- Develop, curate, and promote necessary standards.
- Establish and maintain stable, integrated, and centralized data resources.
- Coordinate public and private contributions.
- Support education and training to ensure a well-informed, well-trained, and diverse workforce.
- Examine and address regulatory and policy challenges.
- Interface with the international community of scientists and funding agencies to identify areas for collaboration and facilitate the sharing of information that may be critical to the overall well-being of humanity and the planet.
The following roadmap provides a general timeline for achieving milestones that the committee has identified as important steps for the implementation of a large-scale, coordinated effort for epitranscriptomics. The actors identified below are suggestions, based on the conclusions and recommendations.
BROADER IMPACT
This report charts a path forward for sequencing RNA and its modifications and presents guidelines that will foster the technology and infrastructure needed to enable, for any cell type or organism, the complete end-to-end sequencing of its epitranscriptome. Investing time, effort, and money in developing the capabilities to determine any epitranscriptome will not only accelerate the understanding of the role that RNA modifications play in living systems and the molecular underpinnings for disease but will also facilitate the application of this knowledge to provide RNA-based solutions to issues in the fields of health and medicine, agriculture, synthetic biology, nanotechnology, environmental science, and beyond. If the information in this report is carefully considered and the recommendations implemented, the committee envisions a day when there will be higher crop yields, affordable treatments for many more human diseases, and vaccines for any infectious disease.
REFERENCES
Barbieri, I., and T. Kouzarides. 2020. “Role of RNA modifications in cancer.” Nature Reviews Cancer 20 (6): 303–322. https://doi.org/10.1038/s41568-020-0253-2.
White House. 2023. Bold goals for U.S. biotechnology and biomanufacturing: Harnessing research and development to further societal goals. https://www.whitehouse.gov/wp-content/uploads/2023/03/Bold-Goals-for-U.S.-Biotechnology-and-Biomanufacturing-Harnessing-Research-and-Development-To-Further-Societal-Goals-FINAL.pdf (accessed March 11, 2024).
Cox, D. B. T., J. S. Gootenberg, O. O. Abudayyeh, B. Franklin, M. J. Kellner, J. Joung, and F. Zhang. 2017. “RNA editing with CRISPR-Cas13.” Science 358 (6366): 1019–1027. https://doi.org/10.1126/science.aaq0180.
de la Torre, D., and J. W. Chin. 2021. “Reprogramming the genetic code.” Nature Reviews Genetics 22 (3): 169–184. https://doi.org/10.1038/s41576-020-00307-7.
Delaunay, S., M. Helm, and M. Frye. 2023. “RNA modifications in physiology and disease: Towards clinical applications.” Nature Reviews Genetics. https://doi.org/10.1038/s41576-023-00645-2.
Elkhalifa, D., M. Rayan, A. T. Negmeldin, A. Elhissi, and A. Khalil. 2022. “Chemically modified mRNA beyond COVID-19: Potential preventive and therapeutic applications for targeting chronic diseases.” Biomedicine & Pharmacotherapy 145: 112385. https://doi.org/10.1016/j.biopha.2021.112385.
Flamand, M. N., M. Tegowski, and K. D. Meyer. 2023. “The proteins of mRNA modification: Writers, readers, and erasers.” Annual Review of Biochemistry 92: 145–173. https://doi.org/10.1146/annurev-biochem-052521-035330.
Gilbert, W. V., and S. Nachtergaele. 2023. “mRNA Regulation by RNA Modifications.” Annual Review of Biochemistry 92 (1): 175-198. https://doi.org/10.1146/annurev-biochem-052521-035949.
Leung, D. W., and G. K. Amarasinghe. 2016. “When your cap matters: Structural insights into self vs non-self recognition of 5’ RNA by immunomodulatory host proteins.” Current Opinion in Structural Biology 36: 133–141. https://doi.org/10.1016/j.sbi.2016.02.001.
Li, N., H. Hui, B. Bray, G. M. Gonzalez, M. Zeller, K. G. Anderson, R. Knight, D. Smith, Y. Wang, A. F. Carlin, and T. M. Rana. 2021. “METTL3 regulates viral m6A RNA modification and host cell innate immune responses during SARSCoV-2 infection.” Cell Reports 35 (6): 109091. https://doi.org/10.1016/j.celrep.2021.109091.
Lowe, D. 2021. “One lost methyl group = huge amounts of food production.” In the Pipeline. https://www.science.org/content/blog-post/one-lost-methyl-group-=-huge-amounts-food-production (accessed September 15, 2023).
Mei, Y., and X. Wang. 2023. “RNA modification in mRNA cancer vaccines.” Clinical and Experimental Medicine 23: 1917-1931. https://doi.org/10.1007/s10238-023-01020-5. National Science Foundation. 2023. NSF 23-554: Molecular Foundations for Biotechnology (MFB) - Partnerships to Transform Emerging Industries - RNA Tools/Biotechnology: Program Solicitation. https://new.nsf.gov/funding/opportunities/molecular-foundations-biotechnology-mfb/nsf23-554/solicitation. (accessed March 12, 2024).
Nelson, A. 2022. Memorandum for the heads of executive departments and agencies. Developed by the Office of Science and Technology Policy. https://www.whitehouse.gov/wp-content/uploads/2022/08/08-2022-OSTP-Public-access-Memo.pdf (accessed March 12, 2024).
“NIST General Information”. 2008. Last modified March 9, 2022. https://www.nist.gov/director/pao/nist-general-information (accessed March 12, 2024).
“Our Mission”. n.d. https://www.ncbi.nlm.nih.gov/home/about/mission/ (accessed March 12, 2024).
Parsons, M. F., M. F. Allan, S. Li, T. R. Shepherd, S. Ratanalert, K. Zhang, K. M. Pullen, W. Chiu, S. Rouskin, and M. Bathe. 2023. “3D RNA-scaffolded wireframe origami.” Nature Communications 14 (1): 382. https://doi.org/10.1038/s41467-023-36156-1.
Poppleton, E., N. Urbanek, T. Chakraborty, A. Griffo, L. Monari, and K. Göpfrich. 2023. “RNA origami: Design, simulation and application.” RNA Biology 20 (1): 510–524. https://doi.org/10.1080/15476286.2023.2237719.
Roundtree, I. A., M. E. Evans, T. Pan, and C. He. 2017. “Dynamic RNA modifications in gene expression regulation.” Cell 169 (7): 1187–1200. https://doi.org/10.1016/j.cell.2017.05.045.
Sahin, U., P. Oehm, E. Derhovanessian, R. A. Jabulowsky, M. Vormehr, M. Gold, D. Maurus, D. Schwarck-Kokarakis, A. N. Kuhn, T. Omokoko, L. M. Kranz, M. Diken, S. Kreiter, H. Haas, S. Attig, R. Rae, K. Cuk, A. Kemmer-Brück, A. Breitkreuz, C. Tolliver, J. Caspar, J. Quinkhardt, L. Hebich, M. Stein, A. Hohberger, I. Vogler, I. Liebig, S. Renken, J. Sikorski, M. Leierer, V. Müller, H. Mitzel-Rink, M. Miederer, C. Huber, S. Grabbe, J. Utikal, A. Pinter, R. Kaufmann, J. C. Hassel, C. Loquai, and Ö. Türeci. 2020. “An RNA vaccine drives immunity in checkpoint-inhibitor-treated melanoma.” Nature 585 (7823): 107–112. https://doi.org/10.1038/s41586-020-2537-9.
Shi, H., P. Chai, R. Jia, and X. Fan. 2020. “Novel insight into the regulatory roles of diverse RNA modifications: Redefining the bridge between transcription and translation.” Molecular Cancer 19 (1): 78. https://doi.org/10.1186/s12943-020-01194-6.
Stojković, V., and D. G. Fujimori. 2017. “Mutations in RNA methylating enzymes in disease.” Curr Opin Chem Biol 41: 20-27. https://doi.org/10.1016/j.cbpa.2017.10.002.
UNESCO (United Nations Educational, Scientific and Cultural Organization). 2023 “Recommendation on open science.”. Last modified September 21, 2023. https://www.unesco.org/en/open-science/about (accessed October 4, 2023).
Winkle, M., S. M. El-Daly, M. Fabbri, and G. A. Calin. 2021. “Noncoding RNA therapeutics — Challenges and potential solutions.” Nature Reviews Drug Discovery 20 (8): 629–651. https://doi.org/10.1038/s41573-021-00219-z.
Yanas, A., and K. F. Liu. 2019. “RNA modifications and the link to human disease.” Methods in Enzymology 626: 133–146. https://doi.org/10.1016/bs.mie.2019.08.003.
Yu, Q., S. Liu, L. Yu, Y. Xiao, S. Zhang, X. Wang, Y. Xu, H. Yu, Y. Li, J. Yang, J. Tang, H.-C. Duan, L.-H. Wei, H. Zhang, J. Wei, Q. Tang, C. Wang, W. Zhang, Y. Wang, P. Song, Q. Lu, W. Zhang, S. Dong, B. Song, C. He, and G. Jia. 2021. “RNA demethylation increases the yield and biomass of rice and potato plants in field trials.” Nature Biotechnology 39 (12): 1581–1588. https://doi.org/10.1038/s41587-021-00982-9.
This page intentionally left blank.





















