Considerations for Returning Individual Genomic Results from Population-Based Surveys: Focus on the National Health and Nutrition Examination Survey: Proceedings of a Workshop (2023)
Chapter: 3 Return of Clinically Actionable Genetic Results (Session 2)
3
Return of Clinically Actionable Genetic Results (Session 2)
This chapter summarizes the presentations and discussion in Session 2, which took place on December 2, 2022, and focused on considerations when returning clinically actionable genetic results. Jeffrey R. Botkin, University of Utah, moderated the session.1
WHAT DATA IS BEING GENERATED?: IMPLICATIONS FOR RETURNING RESULTS
Available Genomic Assays and Their Relative Performance
Matthew Lebo, chief laboratory director at the Laboratory for Molecular Medicine within Mass General Brigham Personalized Medicine, director of bioinformatics for Mass General Brigham Personalized Medicine, associate professor of pathology at Harvard Medical School, and associate member of the Broad Institute of MIT and Harvard, gave the first presentation in this session, which addressed the bioinformatic component of the process of returning genomic results to research participants. Lebo began by reviewing five of the most common genomic assays relevant to the National Health and Nutrition Examination Survey (NHANES). Additionally, the relative
___________________
1 Video recordings of the presentations and discussions, along with copies of the presenters’ slides for Session 2, are available online at https://www.nationalacademies.org/event/12-02-2022/workshop-on-considerations-for-returning-individual-genomic-results-from-population-based-surveys-focus-on-the-national-health-and-nutrition-examination-survey-day-1-virtual
accuracy of these techniques for specific variant identification is compared in Table 3-1.
- Genotyping arrays typically target 500,000 to 2 million sites across the genome. Lebo emphasized the targeted nature of this type of genomic assay. Specific variants of interest are selected before the experiment, and the results allow the variant to be determined at the site of interest. This assay will not identify novel variants outside of the variants of interest. However, it is possible to reanalyze genotyping array data to infer genotypes at untargeted sites using a process called imputation.2
- Targeted sequencing panels rely on the targeted sequencing of specific genes using primers, which limit the scope of base identification to a set number of genes, anywhere from tens to about a thousand. These panels can be enhanced to target specific clinically relevant genes and sometimes are more reliable when used diagnostically for a specific disease.
- Exome sequencing targets only the coding genome (roughly 2% of the entire genome), but it contains all the genetic information cells need to make RNA and proteins. Lebo noted that exome sequencing sometimes has added clinical relevance and is generally less biased than genotyping arrays and targeted panels.
- Whole-genome sequencing (WGS) is the least biased of the genomic assays and can also capture unique information, such as the mitochondrial genome.
- Finally, Lebo described low-pass WGS. Low-pass WGS differs from a typical WGS assay because of the depth of its coverage. A typical WGS array has a sequencing depth or coverage of 30×, whereas a low-pass genome is typically run with 0.25× to 5× coverage. Lebo mentioned that low-pass WGS is beginning to replace genotyping arrays. It has also been adopted in research settings because it is somewhat more accurate for variant calling and has been used for polygenic risk score prediction and in clinical settings for cytogenetic testing (e.g., copy number variant calling). Furthermore, low-pass WGS is slightly more cost-effective than genotyping arrays when performed at scale.
___________________
2 Post-workshop clarification: Imputation uses genomic data from a reference population to predict an individual’s haplotypes and the associated variants present on those haplotypes. The number of sites it can predict is limited by the reference population, and it is typically more accurate for “common” variants.
TABLE 3-1 Summary of File Types and Relative Accuracy for Various Genomic Assays
| File Types | Common Variants | Rare Variants | Copy Number Variations (CNVs) | Structural Variants |
|---|---|---|---|---|
| Genotyping Array | Good, but limited | Poor and limited | Good, but generally for large events | Poor |
| Targeted Sequencing Panel | Good | Good | Low-resolution, unless specifically assayed | None |
| Whole-Exome Sequencing | Good | Good | OK, low-resolution | Poor |
| Whole-Genome Sequencing (WGS) | Good | Good | Good, but low PPVa for small events | Emerging |
| Low-Pass WGS | Good | Poor | Good for larger events | Unknown |
a PPV = positive predictive value.
NOTE: All sequencing methodologies are using short reads.
SOURCE: Adapted from workshop presentation by Matthew Lebo on December 2, 2022 (slide 7).
The Bioinformatic Pipeline
Lebo walked the audience through the bioinformatic pipeline using the sequencing data pipeline as an example, noting that the process is very similar when analyzing data from genotyping arrays. To move data through the bioinformatic pipeline, one must (a) align data with a reference genome, (b) call or identify the variants in the sample that differ from the reference genome, (c) annotate the variants with useful information (e.g., genetic location, population frequency of the variant), (d) filter down to a subset of variants that are potentially relevant to the research or diagnostic question, (e) interpret variants based on the annotations that were previously generated, and (f) report the results.
Lebo next outlined the types of data files created as genomic information moves through the bioinformatic pipeline. The type and size of these files depend on the type of genomic assay employed (Table 3-2) and other key features of the processed data (e.g., annotations). In general, as one views Table 3-2 from top to bottom, the bases read during the genomic assay increases, and therefore, the file sizes increase. However, Lebo noted two exceptions. First, genotyping arrays generate a larger variant call format (VCF) file than targeted panels because genotyping arrays return all genomic information, even if no variant exists. In contrast, the targeted sequencing panel VCF file will contain data only if a variant was detected. The size of VCF files for genotyping arrays can also increase if imputation is used to identify variants at untargeted sites in the genome (see Table 3-2,
TABLE 3-2 File Types and Amount of Data (in MB) Generated by Various Genomic Assays
| File Types | IDATa | CRAMb | VCFc | Imputed VCF |
|---|---|---|---|---|
| Genotyping Array | 60 | n/a | 20 | 750 |
| Targeted Sequencing Panel | n/a | 500 | 5 | n/a |
| Whole-Exome Sequencing | n/a | 5,000 | 100 | n/a |
| Whole-Genome Sequencing | n/a | 30,000 | 1,000 | n/a |
NOTE: File sizes are represented as megabytes (MB).
a IDAT = intensity data file.
b CRAM = compressed reference-oriented alignment map.
c VCF = variant call format.
SOURCE: Adapted from Matthew Lebo workshop presentation, December 2, 2022.
Imputed VCF) and if there are differences in sequencing depth, panel size, and annotations. Lebo also pointed out the considerable size of the raw data file generated by WGS (30 gigabytes per genome) and emphasized this feature of WGS as an important consideration when sequencing many genomes. Finally, Lebo highlighted that there are some emerging tools to combine VCF files and their annotations into a database, which have the potential for financial and computational savings.
Clustering (Arrays), Alignment (Sequencing), and Variant Calling
Lebo explained that when going through the bioinformatic pipeline, the first step is to identify the variants of interest. This process begins differently for the genotyping array than for sequencing assays. In genotyping arrays, clustering is performed before generating the variant calls. Clustering works well with common variants, where it is easier to distinguish between wild-type, heterozygous, and homozygous genotypes.3 However, the clustering method is not very accurate when studying rare variants. This is one challenge of using genotyping arrays to study rare genetic variations. In contrast, Lebo noted that when performing a sequencing array, the first step in the pipeline is to align the genome and call variants. Lebo emphasized the importance of these steps, given that the alignment tool and the variant callers will affect the accuracy. Another reason for variation in research findings is the availability and use of different reference genomes used by the bioinformatics community. Lebo highlighted that using different reference genomes (e.g., use of Genome Reference Consortium Human Build 37 [GRCh37] versus GRCh38) can
___________________
3 Post-workshop clarification: Clustering is the process of defining the exact groupings and when an individual would be called with one of the genotypes (e.g., wild-type, heterozygous, homozygous).
affect variant detection and the availability of certain annotations, and therefore, there is a push for bioinformaticians to adopt the updated GRCh38.4 Lebo also warned that variant caller software also varies, and it is often necessary to use multiple variant callers to capture all relevant variant types, such as copy number variations (CNVs), single nucleotide variants, and insertions and deletions. In closing, Lebo shared that even when best practices are followed, certain genomic variants will be missed. It is important to acknowledge this technical challenge when returning genomic data.
Annotation and Filtration
Lebo next described annotation and filtration, the goal of which is to identify pathogenic and likely pathogenic variants. This differs from diagnostic assays, where genes of interest are prioritized based on a preexisting patient phenotype. Numerous software packages exist for annotating and filtering genomic data that can be purchased from bioinformatics companies or found freely available on open-source platforms. Lebo explained that when annotating and filtering results from genomic assays, it is important to balance sensitivity with positive predictive value (PPV). A typical workflow would first identify variants known to be pathogenic using publicly available lists (e.g., Clinical Genome Resource [ClinGen] expert panels5). Next, additional variants would be labeled as “likely to be pathogenic” or “predicted to cause loss of function” using a separate set of publicly available resources (e.g., ClinVar6) or from one’s own laboratory’s work. Lebo explained that once variants of interest are identified, additional software is typically used to determine the approximate population frequency of the variants (e.g., Genome Aggregation Database7). Lebo explained that filtering variations below a specific frequency is useful because doing so can remove some noise generated by the prediction resources.
Variant Review, Interpretation, and Reporting
Next, Lebo emphasized the importance of variant review and interpretation to determine which variants are returnable. When applying filters,
___________________
4 For additional information about this specific example, see https://www.ncbi.nlm.nih.gov/assembly/GCF_000001405.13/
5 ClinGen is a program for developing standard approaches for sharing genomic and phenotypic data. For additional information, see https://www.genome.gov/Funded-Programs-Projects/ClinGen-Clinical-Genome-Resource and https://clinicalgenome.org/
6 ClinVar is a freely accessible, public archive of reports of the relationships among human variations and phenotypes, with supporting evidence. For additional information, see https://www.ncbi.nlm.nih.gov/clinvar/
7 For more information, see https://gnomad.broadinstitute.org/
there is always a balance between sensitivity and PPV. As a result, even well-designed filters will identify variants that are not accurate or actionable. He provided an example from his own research where a genotyping array initially identified many variants after the filtration process, but only 75 percent of those variants were actually returnable. Variants might fail to be reportable for many reasons. Some that Lebo mentioned during his talk included the following: the variant is found to be absent after additional testing (i.e., a false positive); the variant, upon further review, is only weakly associated with pathogenicity; the variant is recessive and being a carrier is not reportable; probe performance was poor during the original genotyping assay; the variant site is multiallelic such that the variation simply represents an alternative wild-type allele, or that the disease associated with the gene is not returnable. He also pointed out that the bioinformatic process identifies variants associated with conditions that are not actionable according to guidelines by the American College of Medical Genetics and Genomics (ACMG) and that having a way to filter these out automatically could be important. Lebo also noted that the bioinformatic research community has established standards for how to interpret different classes of variants, and he emphasized the importance of adherence.8,9,10
Lebo described the process of variant review for genotyping arrays, which differs from review following sequencing assays. Variant review and interpretation are complicated when using genotyping arrays because the ability to detect rare variants accurately is reduced. The sites where inaccuracies occur are often recurrent, making them easier to be wary of. However, potentially meaningful data can be lost because true-positive and false-positive calls often occur at the same site. This happens when sites are multiallelic, with common variants and rare variants at the same site.
___________________
8 Riggs, E. R., Andersen, E. F., Cherry, A. M., Kantarci, S., Kearney, H., Patel, A., Raca, G., Ritter, D. I., South, S. T., Thorland, E. C., Pineda-Alvarez, D., Aradhya, S., and Martin, C. L. (2020). Technical standards for the interpretation and reporting of constitutional copy-number variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen). Genetics in Medicine, 22(2), 245–257. https://doi.org/10.1038/s41436-019-0686-8
9 McCormick, E. M., Lott, M. T., Dulik, M. C., Shen, L., Attimonelli, M., Vitale, O., Karaa, A., Bai, R., Pineda-Alvarez, D. E., Singh, L. N., Stanley, C. M., Wong, S., Bhardwaj, A., Merkurjev, D., Mao, R., Sondheimer, N., Zhang, S., Procaccio, V., Wallace, D. C., Gai, X., … Falk, M. J. (2020). Specifications of the ACMG/AMP standards and guidelines for mitochondrial DNA variant interpretation. Human Mutation, 41(12), 2028–2057. https://doi.org/10.1002/humu.24107
10 Richards, S., Aziz, N., Bale, S., Bick, D., Das, S., Gastier-Foster, J., Grody, W. W., Hegde, M., Lyon, E., Spector, E., Voelkerding, K., Rehm, H. L., and ACMG Laboratory Quality Assurance Committee. (2015). Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genetics in Medicine, 17(5), 405–424. https://doi.org/10.1038/gim.2015.30
Lebo next outlined variant review for sequencing data, which, because it interrogates the whole coding sequence, can identify rare and novel variants, such as rare loss-of-function variants limited to a single family. Comparatively, variant calling for sequencing data usually has high accuracy, but verification may still be necessary to identify false-positive variants. Some bioinformatic processes can help tease out more obvious true positives from false positives; however, these tools will still return some variants of moderate certainty that need to be confirmed before a patient participating in research can appropriately be informed about a potentially important variant. Overall, sequencing provides a more accurate representation of the spectrum of variation across the genome. It can pick up more variants within the interrogated genes than genotyping techniques because of its unbiased nature and because genotyping arrays can miss variants because of missing or poorly performing probes.
Bioinformatic Considerations for Reassessing Previously Collected Raw Genomic Data
Reassessment of data is important because it can identify variants when new evidence associating variants to disease becomes available. Furthermore, more recent algorithms have improved sensitivity and PPVs for several variant types, especially CNVs and structural variants. Re-running old data through updated pipelines can identify both new true positives and new false positives in the original dataset. Lebo mentioned other improvements to bioinformatic pipelines that make reassessment of old data potentially valuable, including updated annotations, transcripts, improved loss-of-function prediction algorithms, and improvements in reference genomes.
Next, Lebo elaborated on the meaningful improvements to reference genomes. He mentioned the Telomere-to-Telomere consortium, which has developed the complete haploid genome (CHM13), which added approximately 200 megabases of genomic content, including regions that were previously difficult to sequence as telomeres and centromeres. This expansion of the reference genome paves the way for new discoveries. Lebo also highlighted the Human Pangenome Reference, which captures a greater genetic diversity across the human species, which has improved the accuracy of variant calling but is still in the early stages of development.
Bioinformatic Considerations for Generating Data Without Returning Results or Choosing Not to Generate Data
Lebo stated that it is uncommon for genomic data to be generated and then for specific potentially returnable variants to be masked or removed
from the dataset. However, it is possible to do so. Instead, researchers typically release unannotated VCF files with or without aggregate data. In this format, it is much more difficult to identify additional actionable variants without someone diving in deeply and trying to identify them.
Although uncommon, choosing not to generate data on rare variants is possible with both genotyping and sequencing arrays using different methods. When using a genotyping array, specific variants could be ignored if removed from the manifest file before variant calling. However, if they are common enough, there could be a risk of imputing these variants later in the pipeline. Choosing not to call rare variants when using sequencing-based assays is more challenging because these methods inherently identify novel variants. However, calling rare variants can potentially be avoided by setting a minimum population frequency (e.g., 5%), such that only variants with a population frequency above a predetermined minimum are retained. However, Lebo warned that this would probably remove potentially important or interesting variants and could complicate downstream analysis.
ETHICAL ARGUMENTS REGARDING THE RETURN OF RESEARCH RESULTS
Yvonne Bombard is a genomics health services researcher and scientist at the Li Ka Shing Knowledge Institute of St. Michael’s Hospital, Unity Health, Toronto, associate professor at the Institute of Health Policy, Management, and Evaluation at the University of Toronto, and director of the Genomics Health Services Research Program at St. Michael’s Hospital. Bombard acknowledged the difficulty presented by the question of returning results to research participants. Bombard laid out two sides of the argument. First, “there is a basic but important recognition that returning individuals’ results to keep them abreast of new knowledge is an ethically desirable and important goal.” But on the other hand, the world’s resources are limited, and the cost of achieving this goal can be onerous, especially for research efforts that are often time-limited and have limited financial resources. Bombard highlighted that this added cost is often not within the scope of research goals; she emphasized the difference between clinical care and research. “The goal of research is not to deliver medically important information necessarily but is to produce new knowledge and to benefit society as a whole. Of course, direct benefit to an individual research participant is wonderful if it occurs, and when it does, but it’s not the primary purpose of the research goals.” She warned of the opportunity cost built into returning results in research settings, where inappropriate use of limited resources can negatively impact research success.
Bombard described a shift in the ethical principles that underpin the argument to return individual results from a more classic personal
focus11 to a more collective one that acknowledges the need to honor the increasing contributions that participants make to the research enterprise. Bombard highlighted some of the principles advanced in this collectively focused model of biomedical ethics, such as mutuality, reciprocity, and citizenry. Bombard defined mutuality as “the sharing of information” and emphasized its unique importance in cascade testing, in which communication among families is critical because of the hereditary nature of most genomic disease risks. Bombard described reciprocity, or the notion of exchange, as exemplified more broadly by the growing recognition that ongoing participation and contribution of research participants is fundamental for recruitment, retention, and data quality. Bombard emphasized that this continued participation is necessary to advance research enterprises and is of particular concern in the context of the NHANES project moving forward. Finally, Bombard described citizenry as relating to the trend in biomedical research toward implementing processes that increasingly engage, promote, and empower patients, the public, and research participants. She described how citizenry, through the involvement of diverse stakeholder groups, affects how biomedical research is conducted (e.g., the questions that will be asked in a survey, how research projects are designed) and is a recognition of the need to democratize the research enterprise.
State of Consensus Around Returning Genomic Results
Over time, major policy decisions have impacted the return of genomic results to individuals (see Figure 3-1). Before 2006, policies did not recommend returning individual genetic research results. However, in 2021 these policies were reversed, and there is now ethical and legal consensus to support the return of individual genomic results. Specifically, Bombard stated that the return of “medically relevant, valid, actionable results found during research to participants is encouraged.” In addition, Bombard noted that both the public and the research enterprise increasingly support the return of individual genomic results, perhaps partly because of increasing respect for and involvement of participants. In this new system, participants are invited to reimagine, along with researchers: How should participants be engaged as partners in research? How should research be conducted? What results should be returned?
___________________
11 Bombard referred to the four principles laid out by Beauchamp and Childress (beneficence, autonomy, maleficence, justice) to describe a classical, individually focused view of biomedical ethics.

NOTES: ACMG = American College of Medical Genetics and Genomics; ASHG = American Society of Human Genetics; CSER/eMERGE = Clinical Sequencing Evidence-generating Research/Electronic Medical Records and Genomics consortia; GA4GH = Global Alliance for Genomics and Health; IRB = Institutional Review Board; NHLBI = National Heart, Lung, and Blood Institute.
SOURCE: Workshop presentation by Yvonne Bombard on December 2, 2022 (slide 4).
Current policies12 emphasize the “need for clear, a priori, ethics-approved protocols for the disclosure of these results that recognize obviously the participants’ consent and preference around the disclosure, and consider laboratory quality assurance standards about the validity of those results.” Bombard underscored an important element in emergent policies: “recognizing that there is need to provide access to appropriate expertise to support the return of results and that returning results is resource intensive and needs to be budgeted a priori.” However, she pointed out that the guidelines do not require ongoing updates and therefore recognize that “researchers do not have a duty to constantly hunt the literature or their databases for variants.”
Bombard described a potentially more challenging policy problem: how and when to return individual genomic results to participants after the initial results have been returned in the situation where variants have been reclassified and reinterpreted. Along with the American Society of Human Genetics (ASHG), Bombard helped to develop a position statement on this topic that has since been endorsed by eight other international societies, including one patient advocate society. These organizations agreed that recontacting research participants is necessary under two circumstances: if the reclassification (a) altered clinical management or (b) is related to the health condition for which a participant had donated their sample to aid in the research of that condition. Bombard highlighted some qualifiers to these recommendations. For example, the participant must have initially consented to the return of results. In addition, Bombard pointed out that it must be logistically feasible for the participant to be identified; she acknowledged the logistical challenges that the NHANES program has experienced in this regard. Finally, Bombard added that a study can only be expected to return results to individuals after initial contact if the study has the resources and active funding to support the recontact.
The Wide Spectrum of the “Actionability” of Genomic Results
Next, Bombard recentered the conversation around actionability, as she noted policy recommendations to return “actionable” results. She drew attention to the fact that “there is a broad spectrum of actionability, which includes benefits to the person or to the research participant, to their family, and to society,” and explained that this includes a spectrum of actions, ranging from clinical to nonclinical actionability. Bombard provided Figure 3-2 to help illustrate this concept. Next, she walked the audience through this spectrum of actionability by delivering a series of real-world examples. In
___________________
12 Bombard specifically referenced statements from the National Academy of Sciences and the Global Alliance for Genomics and Health.
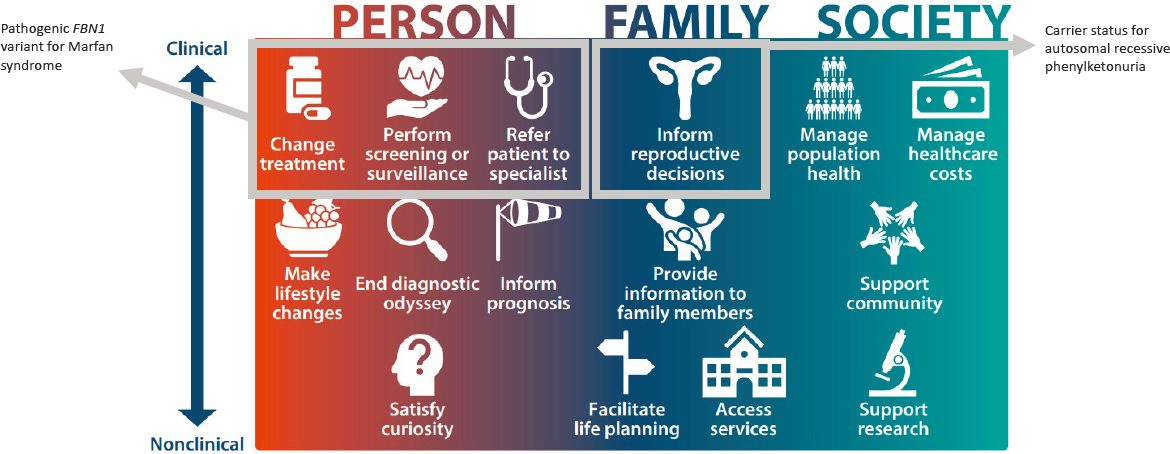
NOTES: Example 1 highlights a scenario in which a patient with a result of a pathogenic FBN1 variant for Marfan syndrome would fit into the actionablity spectrum, leading to a specialist referral, subsequent monitoring for aortic dilation, and an intervention to avoid aortic rupture. Example 2 highlights a scenario in which a patient with a result indicating carrier status for autosomal recessive phenylketonuria, a result which may be considered actionable for a person even if it is not considered actionable by the medical system; this fits in the actionability spectrum by informing personal choices, such as reproductive options, providing information that allows individuals to avoid costly and invasive testing, and providing closure and access to services and supports.
SOURCE: Workshop presentation by Yvonne Bombard on December 2, 2022 (slide 6). Adapted from Goddard, K. A. B., Lee, K., Buchanan, A. H., Powell, B. C., and Hunter, J. E. (2022). Establishing the medical actionability of genomic variants. Annual Review of Genomics and Human Genetics, 23, 173–192. https://doi.org/10.1146/annurev-genom-111021-032401
doing so, Bombard stressed that actionability is interpreted differently by the various groups that make up the biomedical research enterprise and that awareness of these differences in interpretation is essential when developing rules about how to return “actionable” individual genomic results.
Bombard began with an example at the far end of the spectrum, in which actions and interventions (e.g., risk-reducing medications, surgeries, or increased monitoring) have a strong evidence base for improving morbidity and mortality. In her example, giving a patient the result of a pathogenic FBN1 variant for Marfan syndrome can lead to a specialist referral, subsequent monitoring for aortic dilation, and an intervention to avoid aortic rupture. Next, Bombard provided an example in which a result may be considered actionable for a participant even if it is not regarded as actionable in the medical system. For example, a result that “ends a diagnostic odyssey may not impact treatment necessarily but can certainly provide families with the sense of closure and access to services and support groups.” Furthermore, Bombard pointed out that families may find additional benefit if the result allows them to avoid a long series of costly and invasive testing. In still another example, a participant receives carrier status results for a recessive condition like phenylketonuria, which could inform personal choices, such as reproductive options. In one final example, Bombard explained a situation in which someone might see their ability to share their genomic information as having altruistic or societal value. For example, having information about their rare variants could allow participants to sign up for registries like Genome Connect and “allow patients, families, and advocacy groups to improve knowledge globally by sharing their genetic test results and learn about updates as new knowledge evolves over time.” How to reconcile these various ways of actioning or not actioning genetic test results is an ongoing challenge, especially when determining which results are considered actionable to return.
Building on these examples, Bombard emphasized how stakeholders outside the research community have widened the lens of what they consider actionable. She noted that earlier in the workshop, Lebo described a strictly research-focused view of how science determines actionability, citing ClinGen frameworks and the efficacy of potential interventions. However, she pointed out that clinicians and patients can have differing opinions on what should be considered actionable. For example, when Agnes Sabastian, a student in Bombard’s lab, interviewed clinicians, she learned that their view of actionable secondary genetic findings was broader. Clinicians wanted to be able to use genetic information to adapt clinical treatment plans immediately (e.g., reproductive planning, lifestyle changes) and to incorporate the information into their data infrastructure so that these findings could be acted upon later. For example, clinicians were interested in using a pharmacogenomic result that would only be considered actionable
if a patient were to be put on a new medication for which their pharmacogenomic results can inform dosing. Participants broaden the concept of actionability even further, going far beyond what “experts,” whether scientists, clinicians, or members of an expert advisory committee, would consider actionable. Bombard explained that for patients, in addition to clinical management, actionability encompasses actions such as “changing their outlook on life, making plans for the future, and making changes like learning braille, for example, should they find out that they’re at increased risk for hereditary forms of blindness.” If provided with genomic results, participants might feel empowered and employ actions that help them achieve the goal of finding interventions that they are in control of and can improve their quality of life.
Operationalizing and Enabling Actionability
Bombard pointed out that returning results does not ensure action or clinical benefit, and she advocated that research communities create structures to ensure participants can benefit from their putative actionable results. Unfortunately, this is incredibly challenging given the significant inequities in genetic services and health care access—access that is necessary for providing follow-up care for management of high-risk actionable variants. However, she explained, researchers may have a corollary obligation to enable actionability and may “need to put in place workflows, to validate their results, and to ensure patients can receive appropriate followup care, both immediately following return of results, as well as over the life course.” Bombard noted that this is especially pertinent in the case of recording pharmacogenomic results to inform future medication dosing.
Programs such as the BioMe® Biobank Program13 and eMERGE14 have begun to pave the way by developing clinical decision-support tools, such as systems that upload genomic results directly into a patient’s medical record. In Bombard’s own work in Ontario, Canada, she collaborated with a multidisciplinary team of specialists, family doctors, and pharmacogenomic experts to develop a model of care that returns all categories of genetic results (e.g., secondary, incidental findings) and connects the return
___________________
13 Funded by The Charles Bronfman Institute for Personalized Medicine, BioMe is an electronic medical record–linked biobank that enables researchers to rapidly and efficiently conduct genetic, epidemiologic, molecular, and genomic studies on large collections of research specimens linked with medical information. For additional information see https://icahn.mssm.edu/research/ipm/programs/biome-biobank
14 The Electronic Medical Records and Genomics (eMERGE) Network is a National Institutes of Health–organized and funded consortium of U.S. medical research institutions. For additional information see https://www.genome.gov/Funded-Programs-Projects/Electronic-Medical-Records-and-Genomics-Network-eMERGE
of these research results directly to a clinical referral framework. This work aims to serve as a model and can build the evidence base for anticipatory policy and referral frameworks of returning genetic results.
Ethical Considerations of Informed Consent
Bombard highlighted the need to “think about how we meaningfully and feasibly obtain informed consent for research participants, given all the results that may be returned and all the implications that might arise.” A critical workforce shortage in genomics and genetics has made collecting informed consent more challenging, as the number of experts available to support patients is limited. Bombard suggested that this gap in the workforce could be addressed by using digital tools: “The evidence shows that participant-facing tools can be effective at promoting knowledge and decision-making and informed consent.” A recent systematic review found that 84 percent of digital tools have demonstrated favorable, positive impacts on various patient outcomes, including knowledge, understanding, patient decision-making, patient engagement, psychosocial well-being, emotional outcomes, behavioral management changes, and family communication.15
For example, Bombard and her colleagues have developed the Genomics ADvISER tool,16 which aids in decision-making for secondary findings. In a randomized clinical trial, this tool not only was found to be acceptable and highly satisfactory to patients and research participants, but also significantly improved participants’ knowledge and created time savings for genetic counselors. Bombard and her team have since transformed the Genomics ADvISER to be more generally applicable for patient use. “The Genetics ADvISER is meant to be agnostic to any test, patient population, participant population, testing platform, and results.” This tool can be used to help with “up-front informed consent and pre/post genetic counseling, but also the return of results and recontact for updates with variant classifications,” thereby serving as a management and lifelong record for participants.
Disparities in Genomics and Health Care
Bombard has also found that using digital tools to communicate the return of results “promotes patient engagement by enhancing patient-centered care, promoting deliberation, promoting more fulsome dialogue
___________________
15 Lee, W., Shickh, S., Assamad, D., Luca, S., Clausen, M., Somerville, C., Tafler, A., Shaw, A., Hayeems, R., Bombard, Y., and Genetics Navigator Study Team. (2023). Patient-facing digital tools for delivering genetic services: A systematic review. Journal of Medical Genetics, 60(1), 1–10. https://doi.org/10.1136/jmg-2022-108653
16 The Genomics the Genomics ADvISER tool is a Genomics decision AiD about Incidental SEquencing Results (https://research.unityhealth.to/labs/yvonne-bombard/genomics-adviser/).
by the participant, and a wider reflection, or deepened personalization of how they interact with the information.” Bombard asserted that these changes in patient engagement could improve the quality of care by having more participant-centered conversations, which could contribute to the advancement of equity and access in biomedical research. She reminded the audience of the severity of this issue by calling back to Holm’s presentation, in which she described the long-standing biases in genetic databases. Bombard expanded upon this issue by sharing research that shows “a higher prevalence of misdiagnosis from our databases, resulting from increased VUS [variants of unknown significance] rates, false positives, and negatives among individuals of non-European ancestral groups. So not only are there disparities in the data, but disparities in the health care are also well characterized, and these genetic misdiagnoses have the potential to widen these health disparities.”
Bombard pointed out the opportunity for NHANES and other nationally representative samples to diversify databases and close gaps in health disparities. However, she warned that this will depend on the ability to return actionable results in a way that does not exacerbate existing inequities in access to health care. The digital solutions and the actionable variants identified in databases are a product of the data that are entered into those systems, so “if those data are inherently biased, which they are, then these models will unwittingly reinforce those biases, disproportionately disadvantaging those already marginalized in genomics.” Bombard closed by calling upon researchers to “engage, partner with underserved communities, and make sure that our research is representative of broader ancestry groups, the cultural sensitivities of those groups, and make sure that that is done in partnership to avoid reinforcing inequities.”
CURRENT ACMG LIST, PROCESS, AND ALTERNATIVES
Marc Williams, professor and director emeritus of the Geisinger Department of Genomic Health, spoke next about the current ACMG list, including the process used to develop it and its alternatives. Williams explained that 10 years ago and today, most sequencing is done for an indication—meaning there is a valid medical reason to perform the test. For example, a genetic test might be considered when a physician suspects an intellectual disability, multiple congenital anomalies, or some other condition thought to have a genetic etiology. Exome or genome sequencing can provide information about variants in all genes sequenced as part of that assay. This leads to the question: Should analysis of a sequence include genes that are medically important but not related to the indication for testing? Williams explained that current best practice is to report those observations, results, and other findings that may occur during analysis but
are unrelated to the goals of the analysis (e.g., incidental findings) if deemed medically significant.
Williams went on to summarize ACMG’s history on the topic of incidental findings in clinical exome and genome sequencing, starting with the initial policy statement published by ACMG in 2013 that offered a minimum list that initially included 56 genes, as well as several recommendations.17 First, ACMG recommended that laboratories report constitutional mutations18 found in the genes on the minimum list to the ordering clinician, and that the findings should be reported irrespective of the patient’s age. These mutations should only be reported if they were discovered during constitutional or germline analysis, not tumor or somatic analysis. Second, ACMG recommended that only pathogenic or likely pathogenic variants related to the indication be reported. For example, when a clinician finds a variant of uncertain significance in a gene not related to the indication, it should not be reported. This is based on the idea that if an uncertain significance variant is found in a gene related to cancer susceptibility when the indication for the testing was multiple congenital anomalies, the prior probability of this being a condition of interest is very low, meaning that the likelihood that a variant of uncertain significance is a false positive is very high. However, Williams pointed out that this would not be appropriate if a test were ordered for an individual who had a personal or family history suggesting genetic cancer predisposition syndrome that was predicted by the unrelated finding. In this case, even variants of uncertain significance should be reported to allow further analysis. Third, ACMG recommended that the ordering clinician provide pre- and post-test counseling regarding results. Finally, ACMG’s fourth recommendation was against an “opt-out” option, which is consistent with the standard of care that a medically actionable incidental finding should be reported. This standard was considered generally acceptable medical practice in 2012–2013.
Williams next offered several caveats to the 2013 statement: The statement was made at a time when there were insufficient data on penetrance of variants identified in a non-indication-based setting, and there was therefore very little evidence available related to clinical utility to fully support the recommendations. In addition, the 2013 statement acknowledged that the recommendations reflected the “limitations of current technology” for variant detection and interpretation (p. 569). Williams expanded that the intent was to avoid burdening laboratories with additional assays not
___________________
17 Green, R. C., Berg, J. S., Grody, W. W., Kalia, S. S., Korf, B. R., Martin, C. L., McGuire, A. L., Nussbaum, R. L., O’Daniel, J. M., Ormond, K. E., Rehm, H. L., Watson, M. S., Williams, M. S., Biesecker, L. G., and American College of Medical Genetics and Genomics. (2013). ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genetics in Medicine, 15(7), 565–574. https://doi.org/10.1038/gim.2013.73
18 In 2012–2013, the field was using the word mutations rather than variants.
required for the test indication. For example, standard exome or genome sequencing would not detect a trinucleotide repeat expansion; therefore, the laboratory would not be expected to add a test that would detect these solely for the purpose of a secondary findings analysis. Williams also explained that the 2013 statement did not address sequencing for population screening because that was not a consideration at the time. However, the statement did address the topic of using the list in more general research settings. Here the authors stipulated: “Although we hope that investigators find our process and these recommendations useful in their attempts to design thresholds and lists for the return of genomic findings to research participants, we did not design this list for that purpose.”
Williams said that in 2017, ACMG issued an update to the initial recommendation which included significant changes, such as removing one gene and adding three so that the 2017 ACMG list had 59 genes. The update introduced a nomination process for genes to be added or removed, which incorporated the use of growing evidence of clinical utility. A semiquantitative metric for determining actionability consistent with the approach of the ClinGen actionability working group19 “that includes evidence of clinical interventions that can improve health outcomes” was introduced as part of the nomination process. In addition, the terminology was revised to secondary findings instead of incidental findings to reduce confusion for clinicians and to reflect that the genes are intentionally being analyzed. Finally, the 2017 statement also included an option for opting out of receiving secondary findings for individuals undergoing clinical genomic sequencing, reflecting significant ACMG member feedback that an opt-out option is appropriate in this context.
Williams explained that in 2021, ACMG published a third revision to the list, as well as a standalone methodology publication that explained explicitly and transparently why nominated genes were included or excluded. The 2021 publication described a nomination process for the ACMG list, which automatically includes any gene that the ClinGen actionability working group has evaluated and that achieved a score of ten or more. In addition, the 2021 publications specified that the revised ACMG list is for clinical testing and was not validated for general population screening.
Williams went on to share that ACMG has subsequently established two working groups to specifically address the issue of population screening using sequencing: the Genomic Screening of Asymptomatic Patients Working Group and the Population Screening Working Group. However, he reiterated that working groups focus on clinical testing, not research. “Researchers in consultation with their local IRB [Institutional Review
___________________
19 See https://clinicalgenome.org/working-groups/actionability/
Board] should decide on the appropriateness of return of secondary findings for their study,” quoted Williams.
Williams next commented on why he was asked to talk about the return of secondary findings in a research setting, since he repeatedly said during this talk that the ACMG lists were not designed for research studies. Williams went on to list possible reasons. First, the ACMG secondary findings list is the most visible list of secondary finding genes. It is also defined as a minimum list, which allows discretion to add additional genes based on local contextual factors. Second, the ACMG lists have been used in very high-profile research projects focused on return of genomic results, including the eMERGE network, and the Geisinger MyCode Project. Third, the ACMG uses an explicit and transparent methodology that is reviewed regularly and updated as evidence accrues. Furthermore, laboratories have developed pipelines for analyzing secondary finding genes from the ACMG list, making it relatively easy for them to apply that pipeline in a research context without a lot of additional resources. Finally, the ACMG list is updated annually to reflect the latest knowledge and technologies. The next ACMG list, version 3.2, will be published soon and “is much more dynamic than it had been in the past,” offered Williams.
Williams summarized his view on returning secondary findings by reiterating that “researchers in consultation with the IRB should decide on the appropriateness of the return of secondary findings in their study” and “[Bombard] presented a wonderful context for how that discussion could take place.” In other words, which secondary findings to return, how they are returned, and whether updating is appropriate for longitudinal studies are solely at the discretion of the investigators and the IRB and any sort of consultative methods that they are using, interpreted Williams. The informed consent must address the issues of returning secondary findings and opt-outs. Williams used the Geisinger MyCode project as an example of a project that does not allow opt-outs meaning that participation is contingent on allowing the return of results. “If you don’t want to have findings returned, then we say this is not the research project for you.”
Williams next offered his opinion that provisions for the transition of care from the research project setting to clinical care must be considered in the research setting. “We can’t just give the result to a research participant, say good luck with that. Given what we know at least about the United States health care system, that would be a recipe for disaster,” said Williams. Instead, research studies need to account for the resources needed to allow for that transition. The approach must comply with all relevant rules and regulations (those regarding health genomic information, laboratory testing, etc.).
Williams concluded by reiterating another point made by Bombard and displaying a slide titled, “What do participants want?” along with a
visual of Geisinger’s Engagement Framework, which Williams cocreated with Geisinger’s senior ethicist, Dan Davis. “The reason that our MyCode project includes the return of results is that we heard loud and clear from our participants that this is something that they wanted.” He explained that their notion was to take some of the continuums of engagement represented previously in clinical care and quality improvement and move them to discovery, where the dynamic moves from informing participants about activities to consultancy to including them as advisors and ultimately partners. Williams endorsed having a full partnership with research participants, up to and including funded investigators. He shared his opinion that “this is the model that the field is moving to” and something that should be considered when thinking about the return of secondary findings.
ATTITUDES AND OUTCOMES OF RETURNING RESULTS TO PARTICIPANTS
Julie Sapp works as part of a multidisciplinary research team at the Center for Precision Health Research at the National Human Genome Research Institute (NHGRI), where she draws upon her genetic counseling training and over a decade of behavioral science research experience to promote the integration of genomics into medical practice. The diverse clinical research portfolio of the laboratory and her role as a primary clinician working directly with research participants has positioned her to investigate a varying set of important social and behavioral questions related to clinical genomics and genetic counseling. Sapp reminded the audience that although she is a federal employee, the views expressed here are her own. She reiterated that today’s goal is to review how research participants think about and react to options for learning genomic results or their attitudes toward this prospect and what happens after these individuals learn about their genomic findings or the downstream outcomes of this kind of information.
Sapp prefaced her talk with a few key points. First, she highlighted that a lot of the vast literature addressing attitudes has included research participants, the general public, and specific subpopulations, which seems relevant for the NHANES context because of how recruitment for the survey is done. She reminded the audience that much of the research on what happens to people after they learn about genomic research or genomic data generated as a result of research participation focuses on what people do with actionable genomic information. Finally, Sapp stated that in addition to an extensive literature discussing attitudes, there is also a growing body of research on outcomes, including several systematic reviews, and that her rationale for citing a few specific studies is not because they are the only ones addressing a particular idea, but rather because they offer an exemplar of a particular finding.
Research on attitudes toward the return of results addresses thoughts, opinions, motivations, intentions, and preferences of a diverse set of individuals, research participants, and the general public, said Sapp. This includes parents whose children are enrolled in genomic sequencing studies, patients who enroll in studies that use sequencing technology in hopes of learning more about a particular disease or disorder, and other subpopulations. Sapp echoed a sentiment heard in previous talks that one consistent finding reflected in the literature is people’s overwhelming preference for the return of their results. She shared that in many studies, of the people asked about whether or not they want their results, or if they even want the option for their results, 100 percent say they want all genomic data generated as a result of their participation in the research.
Sapp noted a general trend in the literature that as actionability or perceived actionability increases, there is an increasing preference to receive these types of results. She added, “People want lots of results, they want their genomic information, but when they perceive it to be actionable, they want it even more.” Sapp continued by sharing that actionable findings are not the only kind of genomic data that people want. When results are categorized for them in terms of actionability, as several studies have reported, people will say that they want not only findings perceived as normal but also raw genomic data that has not been annotated or interpreted for them in any meaningful way. In a few studies, research participants will say that if the research team doing the sequencing thinks a finding is interesting, then yes, that’s the kind of information that they would like to have, which Sapp thinks shows a sense of trust in the research team who are performing the sequencing.
Sapp shared that another consistent finding across various studies and contexts is that a very small number of people want no genomic data or are only interested in what are considered primary findings—those that relate to the specific disorder under investigation, for example. Sapp pointed to research demonstrating that when looking at the underlying values and beliefs that motivate people to have these generally very positive attitudes, the most frequently cited value is that people perceive genomic data generated from research to be empowering in some way. “They want to use it to prevent disease in themselves, or they think it might be of benefit for family members as well,” Sapp offered. “And so direct quotes from some of these studies will use language like ‘on the one hand, I recognize that this might be a burden, yet, on the other hand, I still really want to know because it could be a benefit to me’ … and this is very common when we ascertain the underlying values and beliefs behind these attitudes,” she said.
Sapp explained that not all of the rationales that people cite have anything to do with health. Research has suggested that people join these studies and want their results just because they are curious or because
they think that it might help them understand themselves or their ancestry better, said Sapp. However, even as people explain their rationale for these generally positive attitudes, they frequently cite concerns such as privacy, the potential for the information to pose a burden to them in some way, or potentially adverse effects on their life or familial relationships. Sapp stated that those concerns do not generally override this preference for return.
Sapp transitioned next to the ideas of ownership and entitlement, which Bombard and Williams also touched on. Sapp shared that research participants perceive their genomic data as something that belongs to them inherently, and they want control over it. Sapp reported that people get uncomfortable when confronted with the idea that a third party might be the one to determine what’s in their best interests. People invoke ideas such as “the right to know” or a sense of autonomy when they think about who ought to be making decisions about what findings they should receive or if they should receive findings at all. Sapp shared that when confronted with ideas about how a third party might be deliberating or restricting what kinds of results to return, this is viewed by research participants as paternalistic.
Sapp added that there seems to be an increasing comfort with broad consent in the sense that participants do not want to be reapproached to reascertain their preference for receiving findings after they have given initial consent. She said, “They’re fine with just agreeing to this once and then being recontacted as needed as new findings come up.” Sapp highlighted one research study that specifically focused on African Americans participating in a large-scale sequencing study of healthy individuals. These participants appreciated that results would be returned according to a plan that was specified at the outset when they initially agreed to be in the study, and they thought that this was a way to foster and continue engagement.
Sapp reiterated that when participants are in a position to receive this information, it ought to be accompanied by genetic counseling in some way so that individuals have an appreciation for what their next steps might be after they learn their information. While some studies suggest a slight preference for being communicated face to face, that finding is not universal.
Sapp stated that very little is known about the minority of individuals who refuse to receive actionable variants. Studies investigating reasons for refusals often pose hypothetical scenarios to individuals, asking them what types of things they would want to know about if they were to join a hypothetical study where they may have the opportunity to learn actionable genetic findings. Sapp highlighted one large biobank study that, upon adding genomic sequencing to the protocol, recontacted people who initially refused secondary findings and asked them to opt in or out of receiving secondary findings. Of 8,843 enrolled, 165 (1.8%) refused secondary findings.
When recontacted, half of those initial refusers changed their minds during the recontact interview and said they did want to receive actionable findings after all. Sapp shared that most of those recontacted believed that they had agreed to receive the findings in the first place and did not understand why their preference was noted as having opted out. Sapp felt that this finding speaks to the need for robust procedures around informed consent and finding a way to explain effectively what is meant by actionable genomic data.
Sapp turned from the topic of attitudes about returning results to outcomes after receiving results. She reflected that while there has been a robust debate in the literature for several years about returning actionable genomic data to research participants and potentially adverse psychological outcomes for patients, she does not think there is a large evidence base to support this notion. Citing multiple studies, Sapp explained that when asked about their emotions after receiving an actionable finding, the emotions expressed ranged from surprise to relief to gratitude to sadness. When explicitly asked about regret, a small minority (0–5%) of recipients said they regretted their decision to opt in to receiving actionable results.
Depression and anxiety have not been investigated in many studies, but when they have been, this appears to be unchanged (i.e., returning actionable variants does not seem to change people’s levels of depression or anxiety). In fact, in one study, some of the recipients of secondary findings or actionable findings, when recontacted to ask them more about their experience, had completely forgotten their finding. They could not remember receiving it, said Sapp. “I don’t think that there’s a huge evidence base for this causing widespread adverse psychological outcomes.”
High numbers of recipients of secondary findings report sharing their results with their relatives, especially those close to them, reported Sapp. She explained that people commonly report disclosing their results to physicians like primary care providers and specialists—around 75 or 80 percent in studies that have investigated this. However, she reflected that sharing secondary findings is not universal and pointed out that “we have a ways to go there.”
Sapp said that when actionable results are returned to people, they are often told to “change their screening and surveillance behavior.” However, the degree to which individuals adhere to the recommendations varies widely, said Sapp. Most recipients of secondary findings and actionable variants receive genetic counseling and are referred to specialists. The few studies that have investigated costs or impact on the health care systems or for people individually show that costs might be lower than expected.
Sapp shared that the literature contains reports of people receiving lifesaving treatment or life-prolonging treatment after receipt of actionable genomic information. Examples include:
- medications to avoid or to reduce the changes of malignant arrhythmias,
- targeted therapies for familial hypercholesterolemia that are picked up not clinically but through return of a genomic variant,
- early identification of malignant lesions by imaging, and
- people undergoing prophylactic surgeries to reduce cancer risks.
Sapp concluded by reminding the audience that “actionable findings recipients are actually a very understudied population.” She shared that her group undertook a systematic review of the literature, outlining how they might want this process to evolve. She used Figure 3-3 to visualize the results, explaining that the size of the blue circles is proportional to the number of reports found in the literature with reports mapped to different parts of the return-of-results process.
She highlighted that there are many more reports in the literature on how secondary findings (SFs) are disclosed and what recommendations are made (Figure 3-3: SFs disclosed and recommendations made, 647 reports), compared to the downstream outcomes in the receipt of that information (Figure 3-3: ongoing surveillance for 6 and earlier diagnosis, treatment, and prevention for 16). Sapp concluded that health outcome data are largely lacking in this population of individuals, sharing that her team was able to find evidence of ongoing surveillance or earlier diagnosis, treatment, or prevention for only 3 percent of the reports.
ISSUES REGARDING DATA REANALYSIS FOR ACTIONABLE VARIANTS
Natasha Strande is an assistant professor in the Department of Genomic Health and in the Autism and Developmental Medicine Institute at Geisinger, and the clinical laboratory director for the MyCode Community Health Initiative. She gave the final talk of this session and spoke about the particularly challenging issues that arise regarding data reanalysis for actionable variants. Strande began by underlining the dynamic nature of genetic knowledge, which encompasses the identification of novel genes and conditions, as well as changes in the understanding of established conditions. Given this constantly evolving landscape, Strande emphasized the importance of continuing to think “about the addition of genes over time and the increased patient population we can be reaching.”
Rate and Directionality of Gene/Variant Reclassifications
Current gene–disease relationships and variant pathogenicity classifications represent the scientific community’s confidence regarding whether a

NOTES: SF = secondary finding. The size of the blue circles is proportional to the number of reports Sapp and colleagues found in the literature.
SOURCES: Workshop presentation by Julie Sapp on December 2, 2022 (slide 18). Adapted from Sapp, J. C., Facio, F. M., Cooper, D., Lewis, K. L., Modlin, E., van der Wees, P., and Biesecker, L. G. (2021). A systematic literature review of disclosure practices and reported outcomes for medically actionable genomic secondary findings. Genetics in Medicine, 23(12), 2260–2269. https://doi.org/10.1038/s41436-021-01295-7
variant causes disease. Building consensus around a gene–disease relationship can take time and is “contingent on the prevalence of the condition and the contribution of a particular gene to the disease.” Still, ClinGen currently has classifications for 1,586 genes, which are subject to change as more information becomes available. Strande acknowledged that variant reclassifications have recently been “very prevalent in the literature.” For example, Strande highlighted several studies that reclassified approximately 5–15 percent of the assessed variants, and one study that found that variant reclassifications affected 22 percent of participants.20 Strande explained that variants are reclassified for several reasons, primarily because of new data availability and “evolving gene-specific variant interpretation guidelines.”
Strande next highlighted the bidirectionality of variant reclassifications: “Confidence can go up or down as we learn more about a specific variant.” She illustrated this point by sharing a few examples from the literature that suggested that, when reclassified, variants are more often downgraded than upgraded. For example, when Harrison and Rehm evaluated the reclassifications of the ClinVar database,21 they found that “the vast majority of the reclassifications were due to downgrades,” with 35 percent changing from VUS to likely benign, and 2.6 percent from likely benign to benign. However, Strande brought attention to this study’s smaller but important number of reclassifications (14.6%) that were upgraded from VUS to likely pathogenic or pathogenic.
Perspectives on Reanalysis from Geisinger’s MyCode Community Health Initiative
Next, Strande shared her experience at Geisinger with respect to updating the MyCode cohort’s disclosure gene list. She explained that Geisinger is an integrated health care system in the central and northeastern Pennsylvania region with “a stable population and long-standing electronic health record data.” Strande described the precision medicine research project known as the MyCode Community Health Initiative as “a biobank that links biospecimens with participant electronic health record data for research purposes.” One aspect of this project involves screening for and confirming variants within exome sequencing data and disclosing actionable findings to participants for use in clinical care. Strande briefly
___________________
20 Machini, K., Ceyhan-Birsoy, O., Azzariti, D. R., Sharma, H., Rossetti, P., Mahanta, L., Hutchinson, L., McLaughlin, H., MedSeq Project, Green, R. C., Lebo, M., and Rehm, H. L. (2019). Analyzing and reanalyzing the genome: Findings from the MedSeq Project. American Journal of Human Genetics, 105(1), 177–188. https://doi.org/10.1016/j.ajhg.2019.05.017
21 Harrison, S. M., and Rehm, H. L. (2019). Is “likely pathogenic” really 90 percent likely? Reclassification data in ClinVar. Genome Medicine, 11(1), 72. https://doi.org/10.1186/s13073-019-0688-9
described the process by which MyCode calls variants of interest. First, MyCode applies several gene and variant filters to the exome data and identifies potentially actionable variants. Next, the genomic results are manually reviewed, and final clinical confirmation is performed before the results are returned to participants. Although the process continues to evolve, the gene list MyCode uses is primarily based “on the criteria outlined in the ACMG secondary findings recommendations.” However, the final decision to return results to participants also includes an evaluation of the gene validity, clinical utility, recommendations on secondary findings that might exist within the Geisinger system, and the availability of local expertise to support disclosure of the result.
Recently MyCode shifted from returning results based on the “ACMG secondary findings version 2.0 list with the addition of hemochromatosis (HFE)” to using the ACMG secondary findings version 3.0 list, which includes 14 new genes. When Geisinger compared the variant detection rates using the version 2.0 and 3.0 lists, they found it increased their ability to detect actionable variants. For example, Strande explained that they saw “a 0.8 percent screen positive increase,” which overall is a 35 percent increase in results. Since Geisinger’s internal MyCode gene list already included HFE, the increases in variant detection were slightly more modest. Strande also highlighted some results that were specific to certain disease domains. For example, she shared that “the collective cancer detection rate increased by 0.11 percent while cardiovascular saw a much larger bump of 0.34 percent.”22
Evolving Policies and Guidance for Genomic Reanalysis
Due to the rapidly evolving nature of gene–disease and variant–disease associations, Strande stressed the importance of reanalyzing genetic data to ensure optimal patient care. Strande described some projects that exist to “standardize curation of this genetic information, ultimately to provide expert-curated information back to the community so that could be integrated into patient care.” The resulting resources provide frameworks for evaluating gene–disease validity and variant pathogenicity. For example, ClinGen has developed a semiquantitative framework that assigns different levels of strength to a gene–disease relationship, ranging from refuted to definitive. Similarly, ACMG has recommended a five-tiered schema for classifying specific genetic variants, ranging from benign to pathogenic, which is informed by multiple data types, including “population data, evidence of
___________________
22 Strande commented that the increased rate of detection for cardiovascular disease was largely due to the addition of the Titan (TTN) gene to the ACMG secondary findings version 3.0 list.
disaggregation, computation predictive data, and functional data.” Strande specifically mentioned that databases, such as ClinVar, NOMAD, and ACMG, provide valuable and updated information about disease-causing variants and inform the variant classification system.
Strande addressed changes to recommendations that were published in 2022 by the ACMG.23 In these updates, ACMG recommended that laboratories have policies in place that address how they will approach reanalysis and outline how reanalysis will occur at the variant level, where “potentially reclassifying any of the variants that were previously reported” might be necessary and at the case level, where “all the different variants in a particular individual” should be reexamined “to see if there are some that should now be reported or not.” ACMG also recommended that laboratories have a protocol for responding to any external request for reanalysis, which might fall outside the realm of their standard reanalysis protocol. Finally, ACMG recommended that laboratory reports clearly state the possibility of changes to variant classifications over time “as a result of evolving genetic information.”
Challenge to Gene and Variant Reclassification and Reanalysis
Throughout her talk, Strande identified some challenges that can accompany gene and variant reclassification and reanalysis. For example, she reiterated that variant gene lists are not static, and that “updates are occurring much more frequently than in the past,” requiring evolving consideration when thinking about reanalysis. In addition, she cautioned that when considering studies that report on reclassification rates (see Rate and Directionality of Gene/Variant Reclassifications) it is important to remember that the reported reclassification rates are “highly contingent on the type of classification and the time frame between that initial classification and the final classification.” She elaborated that one could observe “higher or lower rates depending on the amount of time that has spanned between initial classification and final classification.” Another challenge could arise when U.S. reclassification data are used to measure reclassification in a non-U.S. population. In the example provided by Strande, however, the variant reclassification rate in patients from Singapore fell within the expected
___________________
23 Williams, H. E., Aiyar, L., Dinulos, M. B., Flannery, D., McClure, M. L., Lloyd-Puryear, M. A., Sanghavi, K., Trotter, T. L., Viskochil, D., and ACMG Advocacy and Government Affairs Committee. (2022). Considerations for policymakers for improving health care through telegenetics: A points to consider statement of the American College of Medical Genetics and Genomics (ACMG). Genetics in Medicine, 24(11), 2211–2219. https://doi.org/10.1016/j.gim.2022.07.017
range based on other recent publications that evaluated U.S.-based populations (6.7%).24
Finally, Strande highlighted that changes in variant classification depend on who is interpreting or reanalyzing the data. ACMG is not the only organization that regularly publishes variant classifications and makes recommendations about medical genomic practices. For example, ClinGen regularly updates supplemental guidance from “expert panel groups,” who provide “further specification” of ACMG guidelines related to particular genes and conditions. She noted that inconsistencies in classifications between resources were particularly challenging. She explained that “if you have multiple different groups that are doing variant classifications, you may end up with a situation where one is calling something VUS and another is calling something pathogenic or likely pathogenic” and highlighted this consideration for awareness.
QUESTIONS AND RELFECTIONS FOR SESSION 2
As session moderator, Botkin started the discussion with an issue Holm raised about the ethics of one-time return of results without recontact or reanalysis, and without direct connections to health care providers. Bombard responded first by explaining that from her perspective as a Canadian it is difficult to disentangle access to health care with research activities that border on medical care. She continued by saying, “this is an articulated position across probably a lot of the expert stakeholders, and increasingly among patient and research participants, is that border between clinical care and research is becoming increasingly porous.” Bombard explained that clinically, medical providers discharge patients in genetics and do not expect the geneticists and clinical staff to “chase after patients.” They do now ask patients to keep them abreast of any personal or family history updates, which then leads to questions of recontact on an ad hoc basis. “But for NHANES, I think that’s a larger conversation about resources and a priori design,” said Bombard. She then noted that the ASHG position statement says that if one does not have the resources or mechanisms to trace someone then there is no feasible way to do that.
Williams offered his thoughts next by saying that within the research context one can define how to do research and take things like recontacting participants into account. Acknowledging his limited familiarity with the wants of NHANES participants, Williams postulated that if one was
___________________
24 Chiang, J., Chia, T. H., Yuen, J., Shaw, T., Li, S. T., Binte Ishak, N. D., Chew, E. L., Chong, S. T., Chan, S. H., and Ngeow, J. (2021). Impact of variant reclassification in cancer predisposition genes on clinical care. JCO Precision Oncology, 5, 577–584. https://doi.org/10.1200/PO.20.00399
to ask NHANES participants, they would hear responses like those shared by Bombard and others in this workshop, which is that participants generally want to know about their genetics and “this is something that might enhance the value to us of participating in the study.” Williams then acknowledged that given the repeated cross-sectional nature of NHANES, one must be careful about somehow compromising the data collected and allow for comparisons over time.
Botkin next asked the speakers and audience members to reflect about the ethics of having a full genome sequencing and filtering the results, as discussed by Lebo, such that the ACMG list would be excluded, but all other results could be analyzed. “Can we ethically filter out things that we could identify but choose not to due to the complexities of the … process?” Williams shared that geneticists and variant scientists are applying filters all the time and noted that most of the information in the exome and the genome is being filtered out. “I think the parameters of how you do the filtering is the relevant question, not do you do filtering,” said Williams. When asked by Botkin if it is okay to filter out clinically actionable variants if they are not the focus of the research, Williams replied that he thinks that, with good rationales for saying what a study will and will not look at, combined with being up-front and fully transparent with participants about what will be done, “you can define your research project the way you want to define it.” He emphasized that this approach allows people to make informed decisions about whether they want to participate.
Lebo offered that he was referring to NHANES releasing the data for other people to look at and bioinformatically scrubbing the data of any potential returnable variants, which is akin to what NHANES is doing currently by only allowing for targeted panels that do not include actionable genes. He continued by explaining that the point he was trying to make was that it is technically difficult to do filtering because there are several different lists and those lists change over time. “So if you actively remove something from the data you’re presenting now, there may be something else they want you to remove in like a year or so,” said Lebo.
Sapp added that a research study needs to recognize that if they are going to “wall off and remove” certain parts of the data, then they need to either have a level of certainty that may not be possible to answer the research question in a way that does not live outside of the specified parameters or acknowledge it as a limitation of the dataset. Bombard echoed Williams’s comments and highlighted a finding from Sapp’s talk that participants do not recall what they consent to initially, and their expectations and retention of the information will change over time. She explained that when expectations are set once and only once during the consent process, one must question whether a patient receiving a copy of nonactionable findings understands that they do not have any actionable findings.
The conversation turned next to a question from Holm for Sapp as to whether the studies she presented on have been conducted in more diverse communities. Sapp responded that “a lot of these data have been generated on fairly homogenous majority type of populations, although people have made attempts like some of the work that I cited to try to specifically interrogate how different communities might engage with this information.” Sapp highlighted this as an “urgent priority” and shared that there are efforts under way to better understand how groups like Native populations or non-English speakers approach this information. She speculated that the themes of control and empowerment are likely present in minority communities.
Bombard shared that her team is conducting a systematic review to develop guidance around “how to respectfully and meaningfully engage underserved, racialized, and minoritized groups in genomics and research” that draws on information outside of genomics and health research, which shows a “significant amount of repositioning.” Bombard offered her opinion that more internal reflection about the positionality and privilege held by researchers and clinicians is needed. She highlighted the value she sees in “raising and amplifying of these community partners in the work that we do going forward.” Botkin noted that there may be an opportunity for NHANES to hear what communities want, how they are responding to the information, and the nature of the communication.
Botkin then asked Sapp to share what is known about false reassurance—the extent to which people get results and conclude that they are no longer at risk for certain conditions. Sapp shared that the idea that having a negative secondary findings report equates to a clean genetic bill of health has been investigated with mixed results in terms of how people perceive the information. She noted one study that looked at whether individuals who received negative secondary findings reports made any changes to their health care surveillance plans (e.g., canceling their mammograms, pap smears, and colonoscopies). Bombard offered a corollary consideration because the ways that participants read and misread into what VUS means could also be a source of false reassurance. “I found that patients and participants just read into our grey zone results in the way that they’d like to see those grey zone results reflected in their realities, and that also could lead to false reassurance,” said Bombard.
Kathy Leppig, a speaker from Session 4, offered her opinion that the clinical world provides some information about false reassurance. “For those of us who do a lot of clinical practice, we are seeing reanalysis come on whole-exome sequencing where there are now pathogenic variants that are being reported where there wasn’t even previously VUS,” said Leppig. She explained that clinicians are going through this process right now.
Botkin next steered the discussion toward a discrepancy he observed in the literature between people who are asked the questions around people’s
desire for secondary results when asked, as the data presented by Sapp suggest that “large percentages of people say yes.” He contrasted this against observations in the clinical world where “you have an at-risk family, and testing is available say for a BRCA125 or HNPCC26 or Huntington’s, large percentages of those family members choose not to pursue testing.” Botkin asked for speakers to comment on what is understood about that discrepancy.
Sapp responded first by explaining that this generally positive view from people in the general population reflects that those individuals have not been selected for research simply because “they’re undergoing sequencing in order to complete or solve or resolve a diagnostic odyssey.” She went on to explain that studies examining the preferences of parents of affected children and other affected populations found that respondents find the idea of receiving secondary variants results to be overwhelming. However, Sapp added, “But they don’t all say no, they still sometimes want that information, even when they are coping with complex health care needs.”
Williams offered, “when you have individuals that are at very high risk for carrying a variant, as high as 50 percent risk, uptake of cascade testing is actually quite low, between 10 and 20 percent.” He noted that this phenomenon is found not only in the United States, where there are a lot of barriers to access, but also in places such as the Netherlands that have a national health system with universal coverage and protections that should eliminate the barriers seen in the United States. However, even in such places the uptake of testing is lower than expected. The reasons for this have not been explored.
Leppig explained that her team has an active project with Nora Hendrickson at Kaiser Permanente (KP) Washington that is addressing some parts of the issue. “We did an initial study that figured out that for every KP member you have probably three to four first-degree relatives who are also Kaiser Permanente members.” This project focused on recruiting 60 patients who are getting testing for hereditary cancer and asking them if they wanted us to communicate the results to their family members who are within KP. Leppig shared that “a number of people” said yes and provided the names, phone numbers, and email addresses of their siblings or parents or children. Those family members were contacted about enrolling
___________________
25 BRCA1 is a gene on chromosome 17 that normally helps to suppress cell growth. A person who inherits certain mutations (changes) in a BRCA1 gene has a higher risk of getting breast, ovarian, prostate, and other types of cancer. For more information, see https://www.cancer.gov/publications/dictionaries/cancer-terms/def/brca1
26 Hereditary nonpolyposis colorectal cancer (HNPCC), or Lynch syndrome, is caused by inherited changes (mutations) in genes that affect DNA mismatch repair, a process that fixes mistakes made when DNA is copied. For more information, see https://www.cdc.gov/genomics/disease/colorectal_cancer/lynch.htm
in the study, but there was not a huge uptick for people who want these results disclosed. Leppig shared that they are just finishing the results of the analyses and have been accepted to present the findings at ACMG.
This page intentionally left blank.

































