Considerations for Returning Individual Genomic Results from Population-Based Surveys: Focus on the National Health and Nutrition Examination Survey: Proceedings of a Workshop (2023)
Chapter: 5 Process for the Return of Genetic Results (Session 4)
5
Process for the Return of Genetic Results (Session 4)
This chapter summarizes the presentations and the discussion in Session 4, which took place on December 7, 2022. This session focused on the process for returning genetic results from a number of different contexts. Adam H. Buchanan, Geisinger, moderated the session.1
INFRASTRUCTURE, EXPERTISE, AND OTHER NECESSARY RESOURCES TO COMMUNICATE RESULTS IN A RESEARCH SETTING
Anastasia Wise is director of scientific return to participants and impact for the All of Us Research Program and leads the responsible return of genomic results to participants. Wise began by explaining that she would be talking specifically about the infrastructure support and work around developing communications for the DNA results that are available as part of the All of Us Research Program. Wise explained briefly that All of Us seeks to be able to invite into the program more than 1 million participants who reflect the diversity of the country from across the United States. The program is intended to establish a research resource that today has data available to researchers from more than 372,000 participants, with
___________________
1 Video recordings of the presentations and discussions, along with copies of the presenters’ slides for Session 5, are available online at https://www.nationalacademies.org/event/12-07-2022/workshop-on-considerations-for-returning-individual-genomic-results-from-population-based-surveys-focus-on-the-national-health-and-nutrition-examination-survey-day-2-virtual
over 80 percent of those participants coming from communities that have historically been under-represented in biomedical research and 45 percent from racial and ethnic minority groups.
Infrastructure
Wise started with the topic of participant choice, which she explained was an important core value of the All of Us Research Program. When participants choose to join and enroll in All of Us by creating an online account, they start off with the primary consent, which consents them to participate in the program and have their DNA genotyped and sequenced for use by researchers. From that point forward, participants have choices about how they want to participate in the program and what information they want to share. Participants are given the opportunity to share their electronic health records and are provided an option as to whether or not they want to receive their DNA results. They are also given the opportunity to explicitly indicate whether or not they are interested in hearing more about different types of DNA results that can be provided back to them at later points in time, at which point those participants are recontacted with additional information about the new choices they can make.
Wise explained that participants go through a series of screens called “informing loops” which provide participants with information about the choices they can make.2 Participants can indicate which DNA results they are interested in and which they are not interested in; they can also indicate that they are unsure or they would like to think about it more. DNA results are only generated if and when a participant says yes. Genetic ancestry and traits are returned almost instantaneously. Other results like the hereditary disease risk report can take months to generate.
Overall DNA results are built off both genotyping and sequencing data, said Wise. These genotyping and sequencing data are produced for the program on all members who have consented to the initial consent, but the specific DNA results are produced only for those participants who indicated they want them. Genetic ancestry and traits results are produced using genotyping data. Health-related results are produced from whole-genome sequencing data.
There are two main options available to participants with respect to genetic ancestry and traits, continued Wise. Participants are asked whether or not they are interested in receiving any information about genetic ancestry and traits. Participants who say yes are then provided with the option to be able to view each result separately. The four different traits
___________________
2 Though not stated aloud, Wise’s slides indicated that informing loops provide participants with the ability to change their minds.
that are available are bitter taste perceptions, cilantro preference, earwax type, and lactose intolerance. Participants interested in receiving genetic ancestry results go through a brief series of additional screens that provide them with additional context about the risks and benefits. In addition to the same information provided previously in the consent, the screens also include more detail about the information participants might learn about their genetic ancestry results. For example, reminding them that this is information about genetic ancestry and where their ancestors might have lived hundreds of years ago versus “being necessarily directly aligned with where their parents might have come from or their own personal family stories,” said Wise.
Wise stated that to date, the All of Us Research Program has provided notifications (email, push, and SMS based on participant preference) to over 180,000 participants allowing them to make the choice about whether they are interested in receiving genetic ancestry and traits information. Over 70 percent of those participants have accessed the online portal. Of those, about 88 percent viewed at least one of the traits results and about 97 percent viewed their genetic ancestry results.
Wise shared that the All of Us Research Program has started to offer participants the option of receiving health-related results. Participants have two sets of options they can choose from: (a) Hereditary Disease Risk and (b) Medicine and Your DNA. Participants can make their choice as to whether or not they are interested in receiving one, both, or neither of these reports. The Hereditary Disease Risk report is based on the American College of Medical Genetics and Genomics (ACMG) secondary finding list version 2.0, which has 59 genes and includes information about potentially medically actionable, serious health conditions, such as cancer or heart disease. Participants can make a separate choice about their Medicine and Your DNA report, which includes seven genes that are related to pharmacogenomics and how medications might be used. The reports are then generated according to the participants’ preferences. Once ready, participants are notified that a particular report is completed and ready for them to view. Some participants may receive a notification that asks them to schedule a genetic counseling appointment. Others may be notified that they can view their results in the online portal. Wise emphasized that the DNA results that are provided back to participants as part of the All of Us Research Program, including health-related reports, are research results that need to be confirmed with a clinical test before they are used in care.
Wise next transitioned to showing an example of what the results look like and how participants access them. In addition to receiving and interacting with the results in the portal, participants can also download reports as a PDF, schedule genetic counseling appointments, access information that can be shared with family members, and request to have a PDF copy of the
report faxed to their health care provider. The online portal also has additional information participants can use to learn more about their results.
Supports
Wise explained that while there is no genetic counseling required for genetic ancestry or traits results, All of Us wants to ensure that any participant who wants to reach a genetic counselor can do so. Participants can reach out by phone, email, or chat, which are answered by the Support Center staff who support the entire All of Us Research Program. Support Center staff are provided with general FAQs that get updated periodically. These supports are available in both English and Spanish, and medical interpretation services are also available in more than 200 languages. Questions that cannot be answered by general Support Center staff can be escalated to the program’s Genetic Counseling Resource Call Center, where staff can answer more detailed questions about genetics and genomics, as well as provide genetic counseling for any participants who have specific questions about their DNA results.
All participants with Hereditary Disease Risk or Medicine and Your DNA results are provided an opportunity to schedule an appointment with a genetic counselor. Health care providers can also reach out directly to the Genetic Counseling Resource. These services are available at no cost to participants and a participant can have multiple appointments. Genetic counseling notes are available to participants in the Genomics Platform portal online. As with the general support center, the genetic counseling supports are available in both English and Spanish, and medical interpretation services are also available in over 200 languages.
Wise shared that the online experience was designed to include “tool tips” and other places where participants can click to get more general genetics and genomics information. This information and more are also available in the participant portal that they can access and explore at any time. The learning center provides additional educational content (e.g., text, images, and videos) for participants who want to learn more, including topics that participants can explore.
Communication
Wise outlined the different comprehension and user testing feedback that were collected during the design phase for the DNA results. She explained that it was important to treat participants as partners in the research enterprise beyond simply giving them the choice of what information they want. The All of Us team wanted to ensure that the information being provided to participants was of value to them and could be understood.
It used a series of comprehension tests for the actual information being provided to participants. For example, initial drafts of study materials used the word pharmacogenomics. The comprehension tests revealed how participants were dissecting the words; while they thought that “pharma” referred to medicine and that “gen” had something to do with genes and genetics, some participants were interpreting the “omics” part of this as being related perhaps to economics or something else. Using these results, the All of Us team realized the word pharmacogenomics was not needed to convey the information that it wanted to impart to participants and instead opted for “Medicine and Your DNA,” which the team thought would “be easily understandable across the diverse cohort of the All of Us Research Program,” said Wise.
Wise shared another finding from the pretesting: some participants were overwhelmed when too much information was included on a single screen. This led the team to revise the information such that the main points are displayed prominently on the screens, while more in-depth content, which may include external links, are made available only by clicking on the “tool tips” icon.
A cultural awareness committee made up of multiple different community partners helped review the content to make sure that it was inclusive of all populations. For example, the committee steered the program away from naming male or female organ systems when talking about specific genes. Instead, the program uses language that refers more specifically to risk based on the sex that a participant was assigned at birth, while also providing the context about why the result is potentially important even if it does not align with a participant’s gender identity.
In closing, Wise emphasized that this was intended to be an iterative process. While the team has gotten a lot of information back from participants on the DNA results, it is still collecting feedback. “We intend to continue taking this information and feedback, learning from it, and adjusting our results in order to be able to make them more understandable, easier to use, and of more value back to our participants.”
COMMUNICATION STRATEGIES IN A CLINICAL RESEARCH SETTING
Kathleen Leppig, chief of genetic services at Kaiser Permanente in Washington and a clinical professor in the Division of Medical Genetics at the University of Washington, explained that she would be sharing what was learned from the third phase of the Electronic Medical Records and Genomics network (eMERGE3). Leppig explained that the goal of eMERGE3 was the intentional return of results of clinically actionable variants to the consenting participants and their health care providers from a
curated set of genes and to incorporate those results in the electronic health record.
Providing a map of the United States indicating the location of the eMERGE sites, Leppig shared that eMERGE consists of ten clinical sites, located primarily in the northeast and Midwest regions of the United States, with sites also in Texas and Washington state. Leppig, along with Georgia Wiesner and Alanna Rahm, developed a questionnaire with more than 30 different parameters to determine the process of return of results for each eMERGE site, with Wiesner conducting the interviews with clinicians at each site. Leppig explained that each clinical site developed its own return-of-results process based on the population recruited and institutional requirements. Several sites had more than one process for the return of results; all processes were approved by the local Institutional Review Board.
Leppig emphasized that there were several points that are important to keep in mind in understanding the lessons from eMERGE3. First, one must consider whether a participant who was selected from a biorepository had consented for enrollment prior to or after their DNA sample was submitted to one of the two sequencing laboratories. Another consideration is whether an individual who declined results disclosure would still be considered part of a site’s cohort, explaining that for some sites participants who did not want to get results were eliminated from the cohort, whereas other sites continued to include individuals who declined. A third consideration is whether a site allowed additional opportunities for a participant to opt out of results disclosure. The final consideration centers around the methods used for the disclosure of results to participants and whether that method required participant engagement.
Leppig described one group of clinical sites where the return-of-results process was relatively passive, with participants receiving their results either by mail or an unscheduled phone call. A second group of clinical sites used a process that required participant engagement, such as making a phone call back, or opening up a portal, or making an appointment with a medical geneticist or genetic counselor. Some sites had multiple methods for results disclosure.
Leppig explained that the team found three essential elements that were important when breaking down the return of results: (a) disclosure of results to participants, (b) informing the health care provider, and (c) uploading the results into the electronic health record. For seven of the ten eMERGE3 sites, participant disclosure was the first step of returning results to participants.
Results
Leppig explained that a total of 25,084 participants were recruited from both biorepositories and community organizations. Of the ten clinical sites, two were pediatric and eight were adult. DNA sequencing was performed at two labs: Partners HealthCare Laboratory for Molecular Medicine and the Baylor College of Medicine and Human Genomic Sequencing Center. The eMERGEseq platform had 109 genes and 1,551 variants. The genomic results returned to eMERGE3 participants were a consensus panel of 67 genes and 14 single nucleotide variants, including 58 of the 59 genes listed as actionable in the ACMG list.
All sites were required to return pathogenic or likely pathogenic variants, said Leppig. Six sites also returned other findings, such as variants of uncertain significance. Only one site returned recessive gene “carrier” or heterozygote status. Four sites returned pharmacokinetics results. Finally, six of the ten sites in eMERGE returned null or negative results. Not all sites included genetic counseling in the return of pathogenic and likely pathogenic variants.
Leppig listed three classes of eMERGE3 DNA results that were tabulated and tracked by the coordinating center at Vanderbilt University: (a) the number of participants with pathogenic and likely pathogenic variants (P/LP variants), (b) the number of participants who completed disclosure of results for these disease-associated variants, and (c) the number of participants who had genetic counseling as part of the disclosure process. Of the 25,083 participants in eMERGE3, 94 percent did not have a disclosable variant and 6 percent had a P/LP variant. Looking more closely at the participants with disclosable variants, the variant was not disclosed in 25 percent of the participants where an otherwise disclosable P/LP variant was found. The study found five predominant reasons for nondisclosure. For nearly one-third of the cases, the variant was not eligible for disclosure. This particularly affected the pediatric sites, where individuals would age out of a pediatric protocol and then had to be reconsented as an adult, Leppig added. The study was unable to contact the participant in 20 percent of the cases that were not returned. In 36 percent of the cases the variant was not disclosed because participants passively declined and 9 percent actively declined, meaning that the participants specifically said that they did not want their results.
Leppig underscored that there was great variability across the sites in terms of the proportion of participants who had variant results returned. Five sites returned variant information to at least 90 percent of the participants with a disclosable P/LP variant. In comparison, the two pediatric sites had low disclosures, returning variant information to around 20 percent of the participants with a disclosable P/LP variant.
Participants Opting Out of Disclosure After Being Consented: A Closer Look
By grouping each site into one of two groups—those where genetic counseling was offered but was not embedded into the return of results and those where genetic counseling was embedded—Leppig and colleagues found a striking result. In the sites where genetic counseling was not embedded, 36 percent of the results were returned to participants with genetic counseling. Where it was embedded, 43 percent of results were returned to participants with genetic counseling. Showing the results in a different format, Leppig emphasized that if genetic counseling is embedded in a return-of-results process, 79 percent of participants will receive genetic counseling versus 38 percent where genetic counseling was offered and not embedded. Another interesting finding was that where genetic counseling was offered but not embedded, 92 percent of participants with P/LP variants had return of results versus only 55 percent where genetic counseling was embedded or required yielding a significant difference in whether results could be disclosed to participants.
Leppig summarized the five design elements that were observed to influence the return of results in eMERGE3. The timing of the recruitment and consent process relative to DNA sample submission and sequencing was important, particularly from biorepositories. Sites that required consented participants to have results disclosure as a condition of their enrollment in the study had a higher proportion of participants with completed results disclosure. Sites that included additional opt-out opportunities had fewer consented participants completing their results disclosure. Sites that required genetic counseling had fewer disclosure results. Lastly, methods that required participant engagement were less successful for disclosing results.
Leppig next offered some not-yet-answered questions about participant engagement. What factors influence engagement of participants through the course of a research study? What are the genetic counseling needs for participants engaging in genomic medicine research? And what is the best way to provide genetic counseling to participants? Is there another model other than the traditional in-person visit with a genetic counselor or a medical geneticist to provide the support to participants?
Leppig next shared results from a substudy conducted at the Washington state site to better understand why people refused return of results. The majority of the substudy participants were recruited from a biobank. Of the individuals who passively refused during the main study and were successfully recontacted during the later substudy, 71 percent declined the second offer for the return of results. The most common reasons for declining were that people did not want to know and had concerns about their insurability. “I think one of the things that we learned from that is when people don’t
engage, I think they’re really saying they don’t want their results,” said Leppig.
Leppig listed several unknowns that she felt need to be understood when returning genomic results. How many patients received genetic counseling outside of the planned disclosure process? To what extent did participants who had results disclosed by mail open the mail? Did participants who received their results outside the traditional genetic counseling sessions understand the significance of pathogenic or likely pathogenic variant? Although outside of the scope of eMERGE3, how frequently do relatives who may be at risk have the opportunity for cascade testing? “We know that [cascade testing] most commonly occurs when there is genetic counseling involved and disclosure of results, and that is something that we all feel is very important in genomic testing,” said Leppig. Finally, are there better methods for providing genetic counseling when disclosing genomic results obtained during a research study? Before moving to the next slide, Leppig noted that the All of Us Research Program should be able to shed some light on the last question.
In closing, Leppig displayed some key findings from a paper by Hana Zouk et al. (2022) and elevated the topic of reanalysis of variants as an ongoing concern as the genetics field learns more and grapples with how to incorporate new information into the genetic counseling for research studies.3 She next expressed a desire that she, Weisner, and Rham write a paper focused on what they would do differently if given an opportunity to redo eMERGE3, with Leppig sharing that their goal “would be to develop a toolbox of best practices for returning genomic results in research and clinical studies.” Leppig concluded by sharing the two articles where most of the information she presented can be found4,5 and thanking her colleagues and the many individuals who participated in eMERGE3.
___________________
3 Zouk, H., Yu, W., Oza, A., Hawley, M., Vijay Kumar, P. K., Koch, C., Mahanta, L. M., Harley, J. B., Jarvik, G. P., Karlson, E. W., Leppig, K. A., Myers, M. F., Prows, C. A., Williams, M. S., Weiss, S. T., Lebo, M. S., and Rehm, H. L. (2022). Reanalysis of eMERGE phase III sequence variants in 10,500 participants and infrastructure to support the automated return of knowledge updates. Genetics in Medicine, 24(2), 454–462. https://doi.org/10.1016/j.gim.2021.10.010
4 Wiesner, G. L., Kulchak Rahm, A., Appelbaum, P., Aufox, S., Bland, S. T., Blout, C. L., Christensen, K. D., Chung, W. K., Clayton, E. W., Green, R. C., Harr, M. H., Henrikson, N., Hoell, C., Holm, I. A., Jarvik, G. P., Kullo, I. J., Lammers, P. E., Larson, E. B., Lindor, N. M., Marasa, M., … Leppig, K. A. (2020). Returning results in the genomic era: Initial experiences of the eMERGE Network. Journal of Personalized Medicine, 10(2), 30. https://doi.org/10.3390/jpm10020030
5 Leppig, K. A., Kulchak Rahm, A., Appelbaum, P., Aufox, S., Bland, S. T., Buchanan, A., Christensen, K. D., Chung, W. K., Clayton, E. W., Crosslin, D., Denny, J., DeVange, S., Gordon, A., Green, R. C., Hakonarson, H., Harr, M. H., Henrikson, N., Hoell, C., Holm, I. A., Kullo, I. J., … Wiesner, G. L. (2022). The reckoning: The return of genomic results to 1444 participants across the eMERGE3 Network. Genetics in Medicine, 24(5), 1130–1138. https://doi.org/10.1016/j.gim.2022.01.015
COMMUNICATION STRATEGIES IN A DIRECT-TO-CONSUMER CONTEXTi,ii
Amy Sturm, currently the director of Population Health Genomics at 23andMe, spoke next about how genetic results are communicated to individuals in a direct-to-consumer context, which can be useful for understanding the expectations that the general public may have when receiving genetic results in noncommercial contexts, such as research. Sturm explained that 23andMe offers three different services—Ancestry Service, Health+ Service, and 23andMe+ Membership—which are ordered online by consumers and sent to them via mail for at-home genetic testing. She provided an overview of the many different types of reports that 23andMe returns to consumers and explained that while some of these reports overlap with those returned by the All of Us Research Program, many are unique 23andMe offerings.
Focusing first on the health predispositions reports, Sturm explained that those include some more common Mendelian conditions, as well as conditions where a single genetic variant has a high penetrance and a high risk for a condition such as hereditary breast ovarian cancer syndrome caused by variants in BRCA1 and BRCA2 or familial hypercholesterolemia. She noted that 23andMe is a genotyping platform, not a sequencing platform, which means that they return selected variants in the genes that are associated with those conditions. She pointed out that several reports that get returned to consumers use 23andMe research data from consented research participants to develop polygenic risk scores that can give individuals insights into the likelihood that they may develop a certain condition (e.g., type 2 diabetes, uterine fibroids). Sturm briefly discussed the wellness and carrier status reports offered by 23andMe before highlighting that 23andMe received authorization from the Food and Drug Administration (FDA) to return pharmacogenetic gen-based reports for CYP2C19, SLCO1B1, and DPYD.
Sturm spoke next about the trust and long-term longitudinal engagement the company has with its customers, which can be helpful in thinking through how another return-of-results program might want to engage with its participants. She explained that more than 80 percent of the individuals who purchase a 23andMe kit consent to participate in research vis-à-vis surveys, which is how polygenic risk scores are produced.6 Sturm shared that in 2020, 7 million genotype customers logged in, and 60 percent of customers who bought their kits before 2015 were still logging into the 23andMe system in the year 2020.
Sturm turned to the topic of how 23andMe got to the point of offering FDA-authorized genetic results reports. In 2013, the FDA raised concerns about the analytic and clinical validity of the 23andMe health-related
___________________
6 See https://permalinks.23andme.com/pdf/23_21-PRSMethodology_May2020.pdf
product. 23andMe was required to conduct, among other things, multiple types of user comprehension studies to make sure that the reports and product are understandable and accessible to customers. The result of the conversations between the FDA and 23andMe resulted in the setting of a threshold of at least 90 percent user comprehension for multiple concepts in the reports and the decision that educational modules offered (or in some cases required in order to view certain sensitive reports) to consumers are adequate for over-the-counter use. Sturm highlighted the user comprehension study reports used for each FDA authorization and explained how the company used an iterative development and user testing process composed of quantitative surveys and qualitative studies. In addition to making sure that consumers understood key concepts such as X-linked inheritance, studies compared different platform modalities and found that user comprehension was similar across different devices (e.g., desktop computer, laptop computer, mobile app). Sturm noted that 23andMe complies with stringent FDA regulations because individuals are receiving their results for informational use outside of health care and genetic counseling in order to drive the conversation with an individual’s health care provider.
Sturm summarized several major take-home messages from the user comprehension studies from which she felt anyone developing return-of-results tools, processes, and reports could benefit. First, she said, comprehension research ought to focus on no more than three to five main concepts that are critical for understanding the result because people generally will not recall more than that. When posted with a lot of information, most people do not read everything, so it is important to keep reports as short and succinct as possible. Echoing Wise’s comments, Sturm noted the importance of the information hierarchy and design, including determining up front which results are primary (i.e., those main concepts that need to be front and center in the report and repeated) versus secondary (i.e., those concepts that are important but not critical for the individual to understand their result, which can be found further down in a report or behind a click). Each individual needs to understand the purpose of the test and what the results mean to them; they also must understand the limitations of the testing, including the role of nongenetic factors, said Sturm. She underscored the importance of using white space, illustrations and icons, and colors, so that customers can easily skim and interpret the report; she showed a few examples of specific reports to demonstrate how 23andMe operationalized these principles.7
___________________
7 Sturm’s PowerPoint presentation includes the following example reports: a Genetic Health Risk Report focused on late-onset Alzheimer’s disease, a Medication Insights Report, and a Wellness Report. The presentation is available online at https://www.nationalacademies.org/event/12-07-2022/workshop-on-considerations-for-returning-individual-genomic-results-from-population-based-surveys-focus-on-the-national-health-and-nutrition-examination-survey-day-2-virtual
In closing her presentation, Sturm shared a story to highlight some benefits of returning genetic health risk information, the infrastructure needed to support individuals receiving results, and challenges with health care access after receiving results. The story focused on a 43-year-old male, “Jarrod” (not his real name), who found out through 23andMe genetic testing that he has two variants for a condition that significantly increases the risk of developing colorectal and some other cancers during his lifetime. As a result of receiving this result and reading the recommendations from the report, Jarrod was compelled to see a specialist and receive a colonoscopy much earlier than he had been planning before receiving the genetic-testing results. The initial screening found polyps, and subsequent rounds of testing showed significant increases in the number of polyps. After confirmatory clinical genetic testing, Jarrod ultimately opted into surgery to have his colon and rectum removed. “His physician also shared that given the number of polyps he was generating in such a short amount of time and the number he already had, that had he waited until the age of 45 to 50 to get a colonoscopy, perhaps he would have even had cancer, if not progressive cancer by that time.” Jarrod shared this information with family members who also underwent genetic testing.
iDisclaimer from 23andMe: The 23andMe PGS test includes health predisposition and carrier status reports. Health predisposition reports include both reports that meet FDA requirements for genetic health risks and reports which are based on 23andMe research and have not been reviewed by the FDA. The test uses qualitative genotyping to detect select clinically relevant variants in the genomic DNA of adults from saliva for the purpose of reporting and interpreting genetic health risks and reporting carrier status. It is not intended to diagnose any disease. Your ethnicity may affect the relevance of each report and how your genetic health risk results are interpreted. Each genetic health risk report describes if a person has variants associated with a higher risk of developing a disease, but does not describe a person’s overall risk of developing the disease. The test is not intended to tell you anything about your current state of health, or to be used to make medical decisions, including whether or not you should take a medication, how much of a medication you should take, or determine any treatment. Our carrier status reports can be used to determine carrier status, but cannot determine if you have two copies of any genetic variant. These carrier reports are not intended to tell you anything about your risk for developing a disease in the future, the health of your fetus, or your newborn child’s risk of developing a particular disease later in life. For certain conditions, we provide a single report that includes information on both carrier status and genetic health risk. Warnings & Limitations: The 23andMe PGS Genetic Health Risk Report for BRCA1/BRCA2 (Selected Variants) is indicated for reporting of the 185delAG and 5382insC variants in the BRCA1 gene and the 6174delT variant in the BRCA2 gene. The report describes if a woman is at increased risk of developing breast and ovarian cancer, and if a man is at increased risk of developing breast cancer or may be at increased risk of developing prostate cancer. The three variants included in this report are most common in people of Ashkenazi Jewish descent and do not represent the majority of BRCA1/BRCA2 variants in the general population. The MUTYH-Associated Polyposis Genetic Health Risk Report is indicated for reporting the Y179C and G396D variants in the MUTYH gene and an increased risk for colorectal cancer. The two variants included in this report are most common in people of Northern European
descent. These reports do not include variants in other genes linked to hereditary cancers and the absence of variants included in these reports do not rule out the presence of other genetic variants that may impact cancer risk. The PGS test is not a substitute for visits to a healthcare professional for recommended screenings or appropriate follow-up. Results should be confirmed in a clinical setting before taking any medical action. For important information and limitations regarding each genetic health risk and carrier status report, visit 23andme.com/test-info/
ii23andMe PGS Pharmacogenetics reports: The 23andMe test uses qualitative genotyping to detect 3 variants in the CYP2C19 gene, 2 variants in the DPYD gene and 1 variant in the SLCO1B1 gene in the genomic DNA of adults from saliva for the purpose of reporting and interpreting information about the processing of certain therapeutics to inform discussions with a healthcare professional. It does not describe if a person will or will not respond to a particular therapeutic and does not describe the association between detected variants and any specific therapeutic. The CYP2C19 Pharmacogenetics report provides certain information about variants associated with metabolism of some therapeutics and provides interpretive drug information regarding the potential effect of citalopram and clopidogrel therapy. Results for SLCO1B1 and DPYD and certain CYP2C19 results should be confirmed by an independent genetic test prescribed by your own healthcare provider before taking any medical action. Warning: Test information should not be used to start, stop, or change any course of treatment and does not test for all possible variants that may affect metabolism or protein function. The PGS test is not a substitute for visits to a healthcare professional. Making changes to one’s current regimen can lead to harmful side effects or reduced intended benefits of one’s medication, therefore one should consult with their healthcare professional before taking any medical action. For important information and limitations regarding Pharmacogenetics reports, visit23andme.com/test-info/pharmacogenetics/
SPECIAL ISSUES FOR RETURNS TO PARTICIPANTS IN LOW-RESOURCE COMMUNITIES: LESSONS FROM THE HEALTHY NEVADA PROJECT
Gai Elhanan and Joe Grzymski collaborated to provide an overview of the challenges of returning genomic results in a small community setting, using insights from their work on the Healthy Nevada Project (HNP). Elhanan is a physician in internal medicine and infectious disease and a health care researcher with the Center for Genomic Medicine at the Desert Research Institute and the HNP. Grzymski, who presented to the audience, is the chief scientific officer of Renown Health, the director of the Renown Institute for Health Innovation, and the principal investigator of the HNP.
The HNP is a large-scale population genetics and health determinants study in northern Nevada that performs Exome+ sequencing in laboratories accredited according to Clinical Laboratory Improvement Amendments (CLIA) regulations and the College of American Pathologists (CAP) and in collaboration with Helix.8 The project is organized into a clinical and a
___________________
8 Helix is a private company that conducts population genomics services in CLIA/CAP-certified labs. For more information, see https://www.dri.edu/helix-to-help-expand-healthy-nevada-project/
research component, which currently includes a cohort of 50,000 sequenced Nevadans. Grzymski stated that the goal of the clinical piece of the HNP is “to raise the risk awareness of these autosomal dominant inherited conditions throughout the community.” The clinical component reports on incidental findings on the Centers for Disease Control and Prevention’s (CDC’s) Tier-1 condition list.9 Simultaneously, the research component aims to accelerate research by providing investigators access to valuable health care data (e.g., genomic data, survey data, electronic health records data). By advancing its clinical and research efforts, the HNP seeks to improve clinical care, accelerate biomedical research in Nevada, and drive patient engagement.
Grzymski emphasized that the value of programs such as the HNP and the National Health and Nutrition Examination Survey (NHANES) depends on their ability to improve individual- and system-level clinical care. For example, he offered that when “NHANES was measuring lead, it would have been insufficient to measure lead in children and adults and then do nothing about it. It changed policy and then tracked that downward slope of lead in human tissue, and it actually made an impact.” Grzymski offered that the goal of population genetics health studies, including the HNP and NHANES, should not be limited to returning results but should also aim to monitor and evaluate changes in individual health and inform policies that can improve community health.
Lessons Learned from the Healthy Nevada Project (Version 1.0)
In brief, the first iteration of the HNP (i.e., HNP version 1.0) collected consent and behavioral and social survey data,10 allowed for a “recall” step, and returned genetic results alongside licensed genetic counseling and cascade screening to help individuals understand the implications of a positive autosomal dominant genetic finding. Grzymski highlighted some successes of HNP version 1.0, including a paper that found population screening significantly improved the discovery rate of CDC Tier-1 genetic risks, which were otherwise poorly identified in the general clinical setting.11 In addition, he noted the importance of the recall step in HNP version 1.0, through
___________________
9 CDC Tier-1 conditions include Lynch syndrome, HBAOC (hereditary breast and ovarian cancer) syndrome, and familial hypercholesterolemia.
10 Behavioral and social survey data are used to augment data that can be difficult to extract from electronic health records.
11 Grzymski, J. J., Elhanan, G., Morales Rosado, J. A., Smith, E., Schlauch, K. A., Read, R., Rowan, C., Slotnick, N., Dabe, S., Metcalf, W. J., Lipp, B., Reed, H., Sharma, L., Levin, E., Kao, J., Rashkin, M., Bowes, J., Dunaway, K., Slonim, A., Washington, N., … Lu, J. T. (2020). Population genetic screening efficiently identifies carriers of autosomal dominant diseases. Nature Medicine, 26(8), 1235–1239. https://doi.org/10.1038/s41591-020-0982-5
which participants were informed about potentially beneficial clinical information (e.g., clinical trial availability). This was only possible because, when writing the version 1.0 consent form, HNP staff had the foresight to include language that enabled recontacting participants. HNP version 1.0 also specifically wrote language into the consent form to address Nevada Revised Statute (NRS) 629.181.12 Grzymski highlighted the importance of understanding and complying with all state and local laws, such as NRS 629.181, which might provide individuals with additional rights related to genetic testing beyond federally guaranteed rights. In addition, Grzymski underscored that HNP version 1.0 provided counseling and cascade testing in tandem with positive autosomal dominant genetic results and advised that other programs use the same approach.
Grzymski said that despite these successes, additional research using the HNP data highlighted significant difficulties in translating the return of these results into actions to improve clinical treatment. For example, only 2–3 percent of participants in HNP version 1.0 chose not to consent to the return of results. However, 18 percent of participants were lost to followup (e.g., “did not want to pick up the phone”) or declined their results.13 Furthermore, Grzymski said that 71 percent of participants with positive findings believed they “shared the results with providers, and they felt that the provider and they understood the care path forward.” However, when researchers examined the medical records of these patients, the genetic result was only clearly present in 22 percent and 10 percent of their electronic health records and problem lists, respectively. Finally, this research showed that the genetic diagnoses identified in the HNP did not always lead to care change. Grzymski shared Figure 5-1, an adapted figure from Elhanan et al. (2022),14 which compares individuals’ clinical care after the return of positive genetic results. Participants represented at the top of the figure seem to show that the new genetic finding changed the course of clinical care. In contrast, no change in follow-up care was evident for patients represented at the bottom of the figure. These differences in care exist even though, according to survey results, both groups thought they were “following an action plan” along with their provider based on their returned results (right side of Figure 5-1). Grzymski noted that when NHANES considers why it plans to return results and how it will manage those results, it is essential to reflect on these previous failures to ensure that the clinically actionable findings NHANES discovers are acted upon and benefit patients.
___________________
12 For more information, see https://www.leg.state.nv.us/nrs/nrs-629.html#NRS629Sec181
13 Elhanan, G., Kiser, D., Neveux, I., Dabe, S., Bolze, A., Metcalf, W. J., Lu, J. T., and Grzymski, J. J. (2022). Incomplete penetrance of population-based genetic screening results in electronic health record. Frontiers in Genetics, 13, 866169. https://doi.org/10.3389/fgene.2022.866169
14 Ibid., see Figure 2.
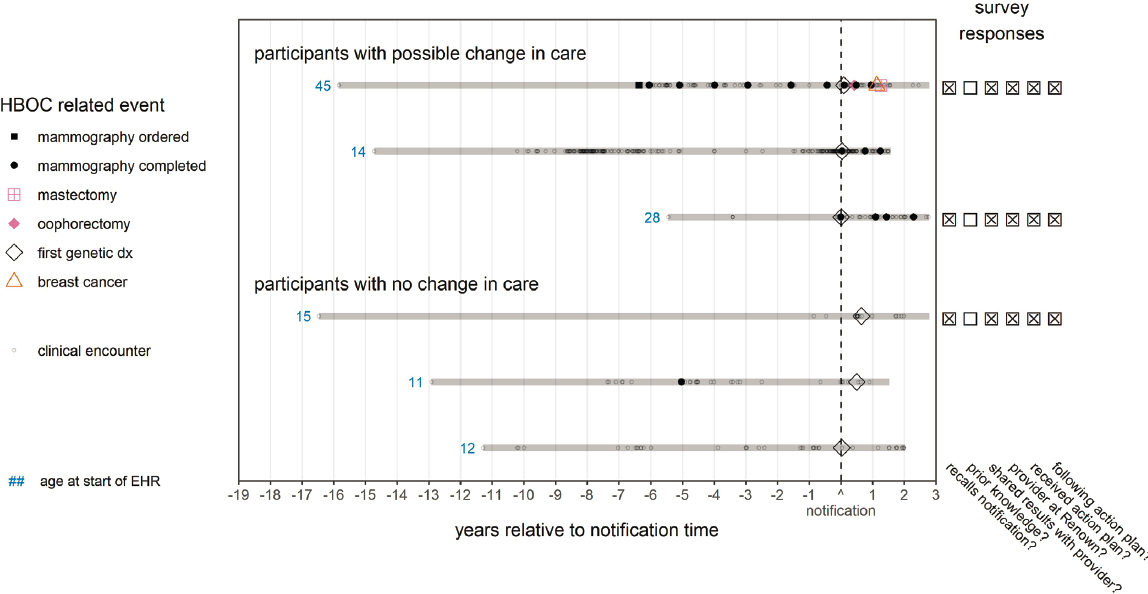
NOTE: HBOC = hereditary breast and ovarian cancer syndrome. Please see the original publication for the full figure and extensive figure notes.
SOURCE: Workshop presentation by Joe Grzymski on December 7, 2022 (slide 6). Adapted from Elhanan, G., Kiser, D., Neveux, I., Dabe, S., Bolze, A., Metcalf, W. J., Lu, J. T., and Grzymski, J. J. (2022). Incomplete penetrance of population-based genetic screening results in electronic health record (Figure 2). Frontiers in Genetics, 13, 866169. https://doi.org/10.3389/fgene.2022.866169
Improvements and Remaining Challenges: The Healthy Nevada Project (Version 2.0)
Informed by those lessons learned from version 1.0, Grzymski spoke next about the evolution of the HNP workflow version 2.0, which is shown below in Figure 5-2. He shared that a root cause analysis by the project team identified four major contributors to their return and implementation failures. First, HNP version 1.0 did not initially enter genetic results directly into the patient’s medical record. Second, HNP version 1.0 returned results using outside genetic counselors with limited ability to coordinate with the patient care team. Third, HNP version 1.0 used off-site research
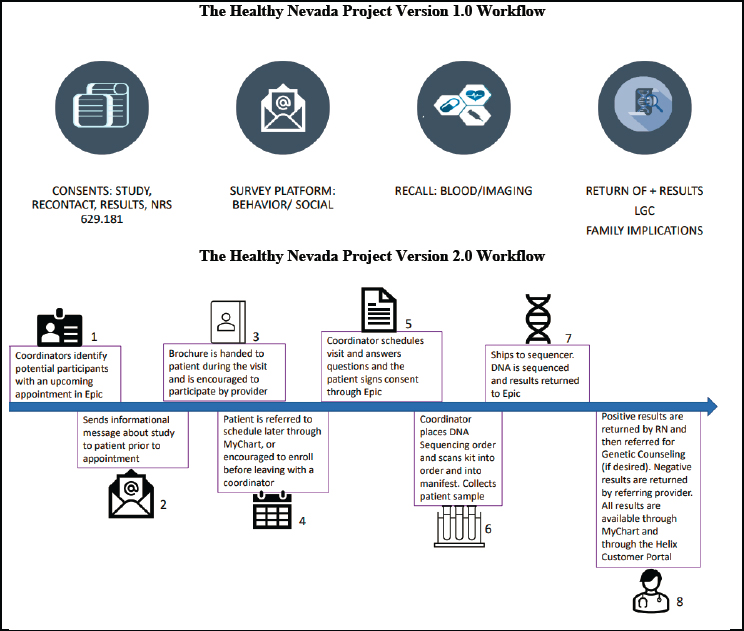
NOTE: LGC = LGC Biotech Technologies (see https://biosearch-cdn.azureedge.net/assetsv6/Genotyping-results-data-from-LGC.pdf); NRS = Nevada Revised Statute (see https://www.leg.state.nv.us/NAC/NAC-629.html); RN = registered nurse.
SOURCE: Workshop presentation by Joe Grzymski on December 7, 2022 (slides 8 and 9).
coordinators to contact patients with positive results. However, these professionals had difficulty reaching participants since their caller ID came up as “Renown” instead of Renown Health. Finally, HNP version 1.0 failed to appreciate the importance of provider education.
Based on this root cause analysis, the HNP implemented several changes to the version 2.0 program. First, to ensure that positive results were included in participants’ medical records, HNP modified the consent form such that individuals could not initially consent to the return of genetic results and then later refuse to receive those results. In HNP version 2.0, all CDC Tier-1 results are returned to patients and entered into the patient’s medical records. Grzymski acknowledged that a major caveat to this recommendation is that some patients, disproportionately in underserved communities, do not have an electronic health care record. In HNP version 2.0, if an individual is not already affiliated with the hospital system, a medical record is created for them. Grzymski also recognized that electronic health records have significant problems. Still, he asserted, including genetic results in electronic health care records is necessary to track individual care to aggregate health care data between hospitals, particularly in a dispersed health care system like northern Nevada.15
A second change was in response to off-site research coordinators’ difficulty contacting participants by phone. HNP had noticed that the return of results was more successful when the call or the contact came from Renown Health. Modifying the off-site research coordination team’s caller ID to read “Renown Health” improved the proportion of phone calls answered. Third, version 2.0 was “more high touch,” with coordinators actively identifying participants, sending potential participants information, and encouraging them to schedule their first HNP appointment and enroll.16 Furthermore, a coordinator maintains contact with the participant to schedule visits, answer questions, and walk them through the consent process. Fourth, in version 2.0, consent is signed through Renown Health’s electronic health care record system, EPIC, and the consent is tied to the patient’s medical record number, “ensuring that the result is returned back into EPIC through an HL7-like interface” which uses an application programming interface to edit and read data from the medical record. A final change that Grzymski highlighted was that in version 2.0, participants with positive results were returned by a trained registered nurse who can also provide genetic counseling where desired, and negative results were
___________________
15 Grzymski emphasized the unique environment of northern Nevada, which contains one health system that provides services for the urban areas of Reno and Tahoe and an expansive area that is “the area of New York, New Jersey, and Connecticut combined.”
16 Grzymski noted that such recruitment is inappropriate in certain settings, such as the emergency room or the intensive care unit.
returned by the referring provider. Grzymski emphasized that providers communicated negative results using “simple catchphrases” to improve understanding. For example, a provider might say, “This just means you don’t have the genetic risk. You revert to normal population-based risk, and here are the appropriate next steps for your care.”
Grzymski shared what he believed to be the new strengths and remaining pitfalls of the HNP version 2.0 workflow. One of the strengths he highlighted is that 100 percent of the results are returned to participants and added to electronic health records. Since patients’ medical records are now flagged with CDC Tier-1 conditions, it has been easier to monitor and evaluate changes in clinical care (e.g., follow-up workflows, order sets, and clinical decision support).
Grzymski noted that returning results within the electronic health record utilizes “the central network of the study” and particularly benefits rural participants who may be underserved or “not have as many clinical touchpoints.” Even if a participant typically visits a rural hospital or health care system, their electronic health record can be updated with the results. However, he noted that special care had to be taken within the consent form to return results through this mechanism since, in his view, NRS 629.181 “requires the consent to deliver those results outside of the specific health system.”
A final strength of the new workflow results from HNP coordinators initiating contact with potential participants, said Grzymski. Active recruitment has generated a more diverse participant population, thus improving the data quality and creating the potential for HNP to use their findings to address historical inequities in the health care system. Pitfalls that remain include difficulty reaching potential participants outside the Renown health care system, clinic-to-clinic variability in provider receptivity of genetic screening that might augment health disparities, and difficulties reaching and providing cascade screening for family members outside of the Renown health care system. In response to these remaining challenges, Grzymski emphasized the need for solutions to improve provider education on the importance of CDC Tier-1 results.
In closing, Grzymski advocated a final time for the critical importance of ensuring that positive autosomal dominant genetic findings, which incur a high risk for increased mortality and morbidity, trigger a change in clinical care for participants and their families. He noted that despite the technical difficulty, HNP spends “a lot of time on cascade screening.” Grzymski highlighted the significant challenge of performing cascade testing meaningfully and informing family members about positive results. To emphasize this point, he shared an example of a couple with two different BRCA variants with many children and family members living within and outside Nevada. In this case, when a sibling who received cascade screening
through HNP asked for help explaining the findings to all other first-degree relatives, HNP had to evaluate each family member’s risk on a case-by-case basis. Grzymski emphasized that returning results requires a commitment to this highly individualized approach and improving clinical care for study participants.
SPECIAL ISSUES FOR RETURNS TO PARTICIPANTS IN LOW-RESOURCE COMMUNITIES: LESSONS FROM THE BIOME PROJECT
Noura Abul-Husn of 23andMe and the Icahn School of Medicine at Mount Sinai gave the final presentation of the session, focusing on the unique challenges of returning results to participants in low-resource communities based on her experience at the BioMe Biobank. She first reminded the audience that large-scale biobanks linking genomic and clinical data are essential to fueling genomic discovery and genomic medicine implementation through screening programs. However, biobank participants are still not representative of global diversity, with many biobanks representing primarily people of European descent. Abul-Husn recognized that there have been some improvements to the diversity within biobanks in general in recent years but that “we’re still a long way away from understanding and representing all of human diversity in our biobank efforts.”
The BioMe Biobank in New York City at the Mount Sinai Health System has more than 70,000 participants and is a good example of a biobank that is representative of a highly diverse patient population, largely consisting of individuals of non-European descent (see Figure 5-3, left panel). At least 55,000 participants have donated exome-sequencing genotype data, and 12,000 participants have donated whole-genome sequencing data. Although BioMe was initially established as a research biobank that did not deliver individual-level results to its participants, that changed in 2018 when a genomic screening program was established to identify, confirm, and return medically actionable genomic results for use in clinical care. Abul-Husn highlighted some of the stakeholders that BioMe worked with during this significant inflection point in their program, including patients, participants, geneticists, nongeneticist domain experts, researchers, health care providers, health care leadership, and payors.
Stakeholder Group 1: BioMe Participants
Abul-Husn explained that the first group BioMe considered were its participants. First, it found that, after BioMe changed its protocol, over 93 percent of participants enrolled to receive genetic results linked to medically
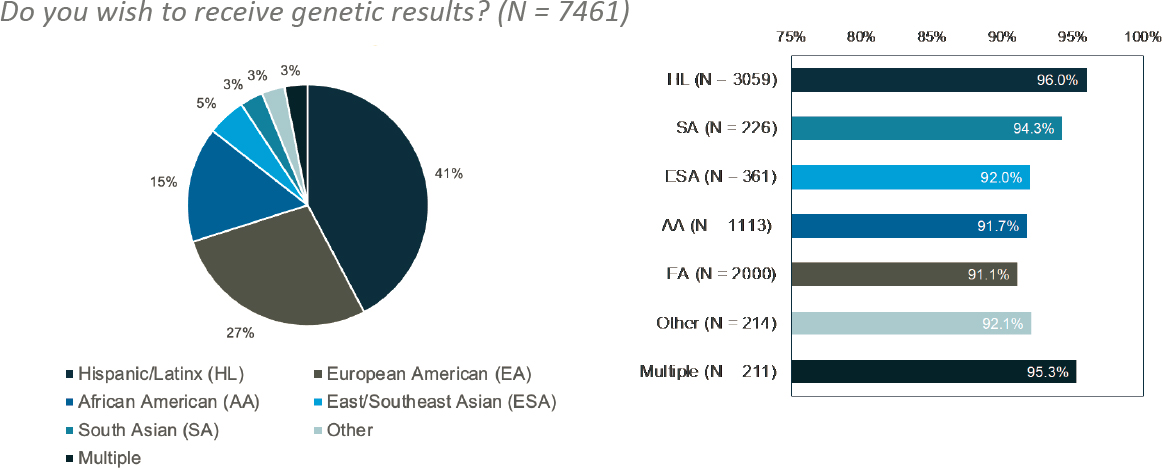
SOURCE: Workshop presentation by Noura Abul-Husn on December 7, 2022 (slide 9). Created using data originally published in Abul-Husn, N. S., Soper, E. R., Braganza, G. T., Rodriguez, J. E., Zeid, N., Cullina, S., Bobo, D., Moscati, A., Merkelson, A., Loos, R. J. F., Cho, J. H., Belbin, G. M., Suckiel, S. A., and Kenny, E. E. (2021). Implementing genomic screening in diverse populations (Figure 3). Genome Medicine, 13, 17. https://doi.org/10.1186/s13073-021-00832-y
actionable conditions. This response varied slightly across demographic groups, but in all groups, at least 90 percent of participants wanted results returned (see Figure 5-3, right panel). Interestingly, it found that participants whose preferred language was Spanish (98%) were more likely than participants whose preferred language was English (93%) to want to receive medically actionable genetic results through BioMe.17 A survey to assess participants’ preferences around how BioMe should implement the return of genetic results asked existing participants how they would like genetic results to be returned. As shown in Figure 5-4, most participants would prefer that a genetic counselor return their results.18 Finally, Abul-Husn mentioned that information about additional questions that were asked of participants could be found in her publication from 2021.19
Abul-Husn shared some of BioMe’s strategies for increasing the accessibility of their genetic screening program, which had a uniquely diverse patient population. These included (a) providing English- and Spanish-speaking clinical research coordinators conducting outreach to communicate about eligibility for receiving results, (b) creating English- and Spanish-language participant-facing materials (e.g., fact sheets), (c) offering point-of-care translation services, (d) providing flexible timing for outreach phone calls (e.g., evenings and weekends), (e) offering flexibility for return-of-results visits (e.g., in-person and telemedicine option), and (f) coordinating in-person return-of-results appointments with existing clinical appointments to reduce transportation and time burdens.
Finally, Abul-Husn gave an overview of key features of the BioMe return-of-results workflow, which was influenced by the participant input and accessibility considerations mentioned above. First, participants can opt out of the return of results. A CLIA-certified and New York State–approved lab clinically confirms all returned results before they are shared with participants. Clinically confirmed results are disclosed to participants in person or through a telemedicine visit with a genetic counselor. Afterward, the results are entered into the patient’s electronic health record and added to their problem list. However, those results that are not disclosed to participants by a genetic counselor do not enter the medical record. Abul-Husn highlighted that much like NHANES, BioMe began as a research initiative not intending to return individual-level genetic results. Therefore, when BioMe changed its protocol, it had a group of 30,000 participants
___________________
17 BioMe has always offered a consent form in Spanish for Spanish-speaking participants.
18 This survey included a description of the types of people who could return results, as listed in Figure 5-4.
19 Abul-Husn, N. S., Soper, E. R., Braganza, G. T., Rodriguez, J. E., Zeid, N., Cullina, S., Bobo, D., Moscati, A., Merkelson, A., Loos, R. J. F., Cho, J. H., Belbin, G. M., Suckiel, S. A., and Kenny, E. E. (2021). Implementing genomic screening in diverse populations. Genome Medicine, 13(1), 17. https://doi.org/10.1186/s13073-021-00832-y
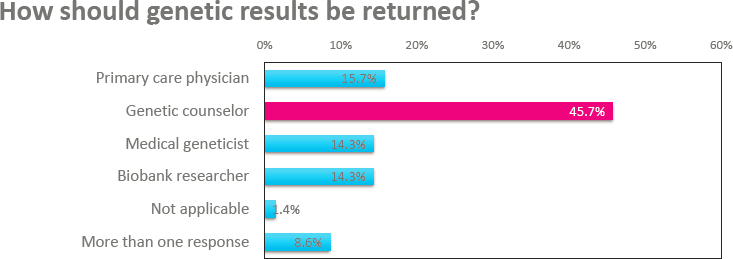
SOURCE: Workshop presentation by Noura Abul-Husn on December 7, 2022 (slide 11). Adapted from Abul-Husn, N. S., Soper, E. R., Braganza, G. T., Rodriguez, J. E., Zeid, N., Cullina, S., Bobo, D., Moscati, A., Merkelson, A., Loos, R. J. F., Cho, J. H., Belbin, G. M., Suckiel, S. A., and Kenny, E. E. (2021). Implementing genomic screening in diverse populations (Figure 1). Genome Medicine, 13, 17. https://doi.org/10.1186/s13073-021-00832-y
who enrolled in the program and had donated exome data but had never consented to the return of results. Therefore, BioMe decided to attempt to recontact the pre-2018 cohort and required that those participants sign an updated consent form to be eligible to receive their results.
Stakeholder Group 2: Geneticists, Researchers, and Health Care Providers
Abul-Husn next explained that BioMe consulted with geneticists and nongeneticist domain experts before it began returning results to determine which genetic findings would be returned to patients. This issue required great care since most research that has generated risk allele gene lists (e.g., ACMG Secondary Findings) has been conducted primarily in individuals of European descent, and the BioMe patient population, in contrast, is very diverse. As a result, BioMe initially limited its gene list to the CDC Tier-1 conditions. However, in continuous collaboration with the relevant stakeholder groups mentioned above, it quickly expanded this list to include TTR, a gene linked to the hereditary transthyretin amyloidosis (hATTR) condition. Before BioMe finalized its decision to include hATTR on its condition list, it established a specialized committee (i.e., the GenomicsFirst Committee) to thoroughly consider the pros and cons of including an additional condition in the program. Abul-Husn described the circumstances that led BioMe to add TTR to its gene list, highlighting the high impact of
hATTR in its patient population and a multidisciplinary centralization of expertise with a targeted interest in hATTR that has the potential to improve clinical care for people properly diagnosed with hATTR.
Abul-Husn expanded on the clinical reasons BioMe added hATTR/TTR to its condition/gene list. She explained that hATTR is highly prevalent, with up to 4 percent of African American individuals and up to 1 percent of Hispanic Latino individuals in the United States carrying the most common variant (i.e., V142I). Furthermore, it is highly penetrant, such that V142I carriers have a 60 percent increased risk of heart failure. Unfortunately, hATTR is underdiagnosed; only 11 percent of people who carry the V142I variant and experience heart failure receive a correct diagnosis. Furthermore, Abul-Husn underscored that the clinical actionability of hATTR played a significant role in BioMe’s decision to add the condition to its program. There are multiple treatment options for patients with hATTR that can delay the progression of amyloidosis, the primary consideration for this disease. However, treatment cannot reverse amyloidosis, suggesting patient outcomes would improve if earlier treatment were provided. Finally, Abul-Husn noted that, historically, hATTR has been an underappreciated genetic condition. However, this is changing as the ACMG Secondary Findings Committee20 added TTR to its gene list of reportable secondary findings in 2022.21
Stakeholder Group 3: Health Care Leadership and Payors
Abul-Husn explained that BioMe, in part, evaluates clinical outcomes because it believes this evidence generation is necessary to increase investment and support from health care leadership and payor stakeholder groups. She expressed her hope that creating this evidence will encourage these stakeholders to support genomic screening. Abul-Husn then shared a few examples of BioMe’s program evaluation that she felt would be relevant to NHANES.
First, she provided an analysis from BioMe relevant to NHANES since it too has a subset of participants who donated DNA before consenting to the return of individual-level results. BioMe compared the return-of-results rates between a subset of people who were part of their first cohort (i.e., participants who enrolled before the 2018 change to begin returning results) and a similar group from the current cohort. Ten percent of the
___________________
20 Abul-Husn is a member of the ACMG Secondary Findings Committee.
21 Miller, D. T., Lee, K., Abul-Husn, N., Amendola, L., Brothers, K. Chung, W., Gollob, M., Gordon, A., Harrison, S., Hershberger, R., Klein, T., Richards, C., Stewart, D., and Martin, C. (2022). ACMG SF v3.1 list for reporting of secondary findings in clinical exome and genome sequencing: A policy statement of the American College of Medical Genetics and Genomics (ACMG). Genetics in Medicine, 24(7) 1407–1414. https://doi.org/10.1016/j.gim.2022.04.006
pre-2018 cohort with positive findings had their results returned to them compared with 31 percent of the new cohort (i.e., who had consented to the return of results at the time of enrollment). Abul-Husn acknowledged this was partially because of differences in recontact rates between these groups. For example, BioMe recontacted 81 percent of post-2018 participants but only 64 percent of pre-2018 participants. To control for this discrepancy, BioMe also looked at the subset of people from each cohort that it successfully recontacted. Still, it found that a much smaller percentage of the contacted individuals from the pre-2018 group consented to receive results. Specifically, only 16 percent of participants enrolled before 2018 received their results. In contrast, 38 percent of participants who enrolled after 2018 and consented to the return of results during enrollment received their results. Based on these data, Abul-Husn concluded that gathering consent to return genetic results at enrollment increases the likelihood of returning the results to participants.
Next, given the problem of hATTR underdiagnosis, Abul-Husn shared research in which BioMe evaluated the clinical outcomes for the first 32 patients who received a positive TTR result through the program. Before BioMe, none of these individuals had genetic testing, nor had been diagnosed with hATTR. Over half of the individuals with TTR V142I had systemic features of hATTR at the time of result disclosure (e.g., heart failure, autonomic neuropathy, peripheral neuropathy).22 Many more had symptoms that may have been related to hATTR but could not be directly linked to the disease. After these individuals received their positive TTR genetic result, 60 percent followed up with a specialist, over half received a cardiac evaluation, and ultimately two individuals (6 percent of those with the TTR V142I variant) were diagnosed with hereditary TTR amyloidosis within 8 months of follow-up.
In closing, Abul-Husn expressed her hope that population-based genomics research, through programs like BioMe and NHANES, will promote health equity through genomics and genomic medicine. In addition, she emphasized that increasing diversity in genomics research is essential to generate knowledge that benefits all populations. Finally, she stated that designing and implementing pilot genomic screening programs tailored to diverse populations is critical, but it is just as important to collect and analyze the data from these programs to inform further research and implementation.
___________________
22 Ten of the patients with the TTR V142I positive result had bilateral carpal tunnel syndrome, which Abul-Husn suggested could be an early indicator of individuals progressing with amyloidosis and hATTR.
QUESTIONS AND REFLECTIONS FOR SESSION 4
Buchanan led and moderated the discussion for this session, which began with a clarification question about how the All of Us Program generates its genetic results. Wise explained that all of its labs’ sequencing data are CLIA certified. However, All of Us has an Investigational Device Exemption (IDE) approval from the FDA for its health-related research results. Therefore, any pathogenic or likely pathogenic findings in the All of Us hereditary diseases research report need only be confirmed with an orthogonal assay before issuing those research results. However, for these research results to be returned to participants and used to modify clinical care, the genetic results must be confirmed again using additional clinical genetic testing methods.
Next, Sturm was asked about the typical turnaround time for 23andMe genetic tests. She explained that since individuals spend their money to receive genetic reports, 23andMe aims to return reports within “a handful of weeks.”
Buchanan next asked the presenters’ thoughts on the importance of returning results through a genetic counselor in research studies. Wise shared that the All of Us Research Program thinks all participants, even those who do not have positive findings, should have access to a genetic counselor by request. However, participants with a pathogenic or likely pathogenic finding on their hereditary disease risk report always receive their results through a genetic counselor. Similarly, Elhanan shared that the Healthy Nevada Project believes a genetic counselor should explain the significance of genetic findings and formulate a clinical action plan as soon as results are returned to participants. However, Elhanan explained that there are not enough medical geneticists to support this. Furthermore, in his experience at the Healthy Nevada Project, primary care physicians are uncomfortable returning genetic results. As a result, he voiced a need for more experts with a higher level of specific knowledge to educate participants about their findings’ significance.
Leppig expressed her concern about the bottleneck that genetic counseling is creating in clinical and research care. Although she stated that she believes genetic counselors are essential to health care and that complex data need to be explained to patients, she suggested that another model might be necessary, depending on the nature of the results, such as when genetic results are not disease associated. Leppig also asserted that normal results do not need to be communicated through a genetic counselor and instead proposed that other providers (e.g., nurses or genetic counseling assistants) should be educated to return results that will not affect clinical care. Sturm agreed with Leppig and Elhanan that relying on genetic counselors to communicate all results is likely not possible at scale. At the same time, she agreed with Wise, stating, “I think genetic counseling doesn’t need
to be a requirement. I think it is important to have available.” In addition, Sturm underlined the importance of giving participants choices throughout population genetic study workflows. She suggested that, just as participants can initially opt out of receiving their genetic results, they should also be able to opt out of genetic counseling. However, she noted that opting out of genetic counseling was only appropriate if a broader infrastructure still exists to follow up on their genomic result and improve their clinical care. For example, genomic results are delivered directly to the electronic health record, enabling medical professionals to act upon them.
Ingrid A. Holm, Harvard Medical School and Boston Children’s Hospital, stated that the Adolescent Brain Cognitive Development Study23 was required to have a genetic counselor return genetic results in order to avoid an IDE; she then asked Wise to expand upon whether and how the FDA stipulates the conditions for the return of positive results under the All of Us Program’s IDE. Wise confirmed that All of Us has an FDA IDE and clarified that the All of Us Research Program staff, rather than health care providers, return results to participants. Because of this possible risk, the All of Us Program had to provide risk mitigation strategies in its application to the FDA, which included the provision of genetic counseling to participants.
___________________
23 For more information, see https://abcdstudy.org/
This page intentionally left blank.



























